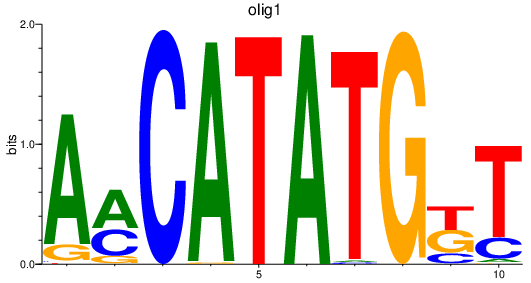
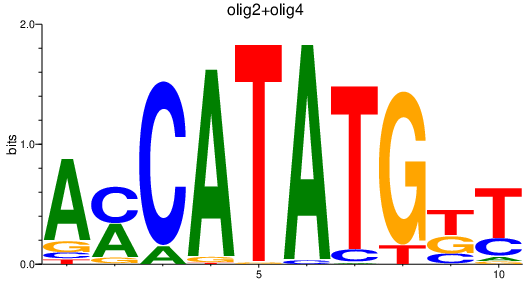
Project
DANIO-CODE
Navigation
Downloads
Results for olig1_olig2+olig4
Z-value: 1.55
Transcription factors associated with olig1_olig2+olig4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
olig1
|
ENSDARG00000040948 | oligodendrocyte transcription factor 1 |
|
olig2
|
ENSDARG00000040946 | oligodendrocyte lineage transcription factor 2 |
|
olig4
|
ENSDARG00000052610 | oligodendrocyte transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| olig2 | dr10_dc_chr9_-_32942783_32942789 | -0.90 | 1.8e-06 | Click! |
| olig4 | dr10_dc_chr13_+_45830454_45830473 | -0.87 | 9.3e-06 | Click! |
Activity profile of olig1_olig2+olig4 motif
Sorted Z-values of olig1_olig2+olig4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of olig1_olig2+olig4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_27394361 | 6.00 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr19_-_27093095 | 5.67 |
|
|
|
| chr2_-_39070943 | 5.54 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr18_-_40718244 | 4.10 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr20_+_34021257 | 3.87 |
ENSDART00000146292
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr9_+_28293831 | 3.85 |
ENSDART00000101338
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr19_+_41419851 | 3.76 |
ENSDART00000138555
ENSDART00000158587 ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr15_+_14656797 | 3.57 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr18_+_284210 | 3.54 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr5_-_27394324 | 3.53 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr9_-_12916555 | 3.49 |
ENSDART00000140691
|
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr21_-_13654658 | 3.24 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr19_-_41820114 | 3.19 |
ENSDART00000038038
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr20_-_22899048 | 2.93 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr4_-_72228164 | 2.88 |
|
|
|
| chr16_-_39317068 | 2.70 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr5_-_14022455 | 2.56 |
ENSDART00000177403
|
CR392037.1
|
ENSDARG00000107645 |
| chr2_-_39071026 | 2.45 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr18_+_18124107 | 2.40 |
ENSDART00000151406
|
BX649612.1
|
ENSDARG00000096293 |
| chr6_-_50731449 | 2.27 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr24_-_23639458 | 2.26 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| KN150663v1_-_3381 | 2.24 |
|
|
|
| chr15_-_5913264 | 2.21 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr18_+_18890626 | 2.19 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr24_-_9549374 | 2.14 |
ENSDART00000093046
|
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr5_-_23741221 | 2.14 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr13_-_51617720 | 2.13 |
ENSDART00000121457
|
lbh
|
limb bud and heart development |
| chr5_+_27659432 | 2.09 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr15_+_14656886 | 2.08 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr20_+_6783996 | 2.07 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr1_-_17000212 | 2.03 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr7_-_24249672 | 1.99 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr13_+_11418060 | 1.96 |
ENSDART00000166908
|
desi2
|
desumoylating isopeptidase 2 |
| chr13_+_11307037 | 1.96 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_+_53366182 | 1.95 |
ENSDART00000162926
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr7_+_9045376 | 1.95 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr5_-_39205454 | 1.92 |
ENSDART00000133231
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr2_-_39070886 | 1.90 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr10_+_9804879 | 1.89 |
|
|
|
| chr20_-_36776382 | 1.88 |
ENSDART00000062908
|
rpl7l1
|
ribosomal protein L7-like 1 |
| chr3_-_7275821 | 1.85 |
ENSDART00000132197
|
BX005085.4
|
ENSDARG00000102658 |
| chr9_-_7235236 | 1.84 |
ENSDART00000092435
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr10_-_33435736 | 1.83 |
ENSDART00000023509
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr10_-_31129270 | 1.83 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr20_-_31349892 | 1.82 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr5_-_54091790 | 1.82 |
ENSDART00000150070
|
ccnb1
|
cyclin B1 |
| chr21_+_11686037 | 1.81 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr14_+_31278584 | 1.81 |
ENSDART00000158875
ENSDART00000026195 |
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr8_-_53625018 | 1.80 |
|
|
|
| chr2_-_7913468 | 1.80 |
ENSDART00000163175
|
dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr22_-_6916274 | 1.79 |
|
|
|
| chr12_+_20685139 | 1.75 |
ENSDART00000153261
|
BX004803.1
|
ENSDARG00000096799 |
| KN150460v1_-_33517 | 1.72 |
|
|
|
| chr13_+_11306913 | 1.71 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_-_17678748 | 1.69 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr10_+_6925373 | 1.69 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr1_+_48770997 | 1.68 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_-_18851683 | 1.68 |
ENSDART00000169180
ENSDART00000006482 |
trpc4apb
|
transient receptor potential cation channel, subfamily C, member 4 associated protein b |
| chr24_-_23639325 | 1.67 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr6_-_22842200 | 1.65 |
ENSDART00000160311
|
sumo2a
|
small ubiquitin-like modifier 2a |
| chr21_+_13291305 | 1.64 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr10_-_31129217 | 1.63 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr21_-_38670059 | 1.62 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr13_+_11417882 | 1.61 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr20_-_31349698 | 1.60 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr19_+_41419985 | 1.59 |
ENSDART00000138555
ENSDART00000158587 ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr17_+_23709781 | 1.59 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr20_+_4034072 | 1.59 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr13_+_11306998 | 1.58 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr6_-_51811024 | 1.58 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr2_+_41909094 | 1.58 |
ENSDART00000179428
|
CABZ01014197.2
|
ENSDARG00000107429 |
| chr7_+_15059782 | 1.58 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr8_-_11806733 | 1.56 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr10_+_15066791 | 1.55 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr12_+_5067663 | 1.54 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr7_+_45747622 | 1.54 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr19_+_10742387 | 1.53 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr8_-_11806698 | 1.53 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr22_-_10861268 | 1.52 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr20_-_34126039 | 1.51 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr24_-_16835352 | 1.51 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr9_+_44327817 | 1.50 |
|
|
|
| chr8_+_7625846 | 1.47 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr5_+_51979524 | 1.46 |
ENSDART00000168317
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr22_-_28276824 | 1.46 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr7_+_38395803 | 1.45 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr20_+_52723371 | 1.43 |
ENSDART00000002787
ENSDART00000158230 |
pycr3
|
pyrroline-5-carboxylate reductase 3 |
| chr13_+_28785104 | 1.42 |
|
|
|
| chr5_-_39598926 | 1.42 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr22_+_1451958 | 1.42 |
|
|
|
| chr24_-_9549419 | 1.40 |
ENSDART00000093046
|
uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_+_24045890 | 1.40 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr13_+_22691356 | 1.39 |
ENSDART00000144094
|
dna2
|
DNA replication helicase/nuclease 2 |
| chr7_+_44463158 | 1.37 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr2_+_10265922 | 1.35 |
ENSDART00000143876
|
cmpk
|
cytidylate kinase |
| chr24_+_36316070 | 1.35 |
ENSDART00000155260
|
rbbp8
|
retinoblastoma binding protein 8 |
| chr1_+_13768098 | 1.33 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr11_+_36117256 | 1.33 |
ENSDART00000141529
|
atxn7l2a
|
ataxin 7-like 2a |
| chr21_+_20350218 | 1.33 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr20_-_23977077 | 1.32 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr17_+_27988842 | 1.31 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr13_+_22567761 | 1.31 |
ENSDART00000137467
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr21_+_13291242 | 1.30 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_-_22099244 | 1.29 |
ENSDART00000161298
|
nono
|
non-POU domain containing, octamer-binding |
| chr12_-_19063761 | 1.29 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| KN150663v1_-_3817 | 1.28 |
|
|
|
| chr25_+_16259634 | 1.28 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr1_-_54586004 | 1.27 |
ENSDART00000152687
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr20_+_23707625 | 1.27 |
|
|
|
| chr2_+_46210971 | 1.27 |
|
|
|
| chr20_+_23707789 | 1.26 |
|
|
|
| chr2_+_46210909 | 1.25 |
|
|
|
| chr22_-_11024649 | 1.25 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr3_+_7877162 | 1.23 |
ENSDART00000057434
ENSDART00000170291 |
hook2
|
hook microtubule-tethering protein 2 |
| chr13_-_51617421 | 1.23 |
ENSDART00000121457
|
lbh
|
limb bud and heart development |
| chr3_+_42667814 | 1.22 |
ENSDART00000172425
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr3_+_25776714 | 1.22 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr12_+_5013049 | 1.21 |
ENSDART00000161548
|
kif22
|
kinesin family member 22 |
| chr2_-_5233961 | 1.21 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr19_+_34263554 | 1.20 |
ENSDART00000161290
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr21_+_20350152 | 1.20 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr11_+_25455772 | 1.20 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr22_-_31089831 | 1.20 |
|
|
|
| chr8_+_44448437 | 1.19 |
ENSDART00000143807
|
mhc1laa
|
major histocompatibility complex class I LAA |
| chr17_+_13803088 | 1.19 |
ENSDART00000176632
|
BX005443.1
|
ENSDARG00000107833 |
| chr15_+_47207251 | 1.19 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_-_15960491 | 1.18 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| KN149790v1_-_67669 | 1.18 |
ENSDART00000163561
|
ENSDARG00000099322
|
ENSDARG00000099322 |
| chr2_+_24046083 | 1.18 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr12_-_20494169 | 1.18 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr20_+_34021116 | 1.18 |
ENSDART00000146292
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr20_-_9135267 | 1.17 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr5_-_14022419 | 1.17 |
ENSDART00000177403
|
CR392037.1
|
ENSDARG00000107645 |
| chr24_-_31394710 | 1.17 |
ENSDART00000169556
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr13_+_28655338 | 1.16 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr3_-_62156743 | 1.16 |
ENSDART00000101870
ENSDART00000175992 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr14_+_35065189 | 1.16 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr1_+_18956542 | 1.15 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr24_-_36721857 | 1.15 |
|
|
|
| chr24_+_16575167 | 1.14 |
|
|
|
| chr6_-_51811058 | 1.14 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr24_+_14451260 | 1.14 |
ENSDART00000137337
ENSDART00000091784 |
thtpa
|
thiamine triphosphatase |
| chr21_+_11685992 | 1.12 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr25_+_4660989 | 1.12 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr24_+_28855974 | 1.11 |
ENSDART00000017427
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr19_-_23665476 | 1.10 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr10_-_14582293 | 1.10 |
ENSDART00000039161
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr3_-_29557642 | 1.10 |
ENSDART00000151679
|
si:ch73-233k15.2
|
si:ch73-233k15.2 |
| chr7_-_24249630 | 1.10 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr25_-_26309873 | 1.09 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr5_-_56640932 | 1.09 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr21_+_20350334 | 1.09 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr8_-_47215582 | 1.09 |
ENSDART00000147606
|
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr20_-_4033876 | 1.09 |
ENSDART00000113184
|
arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr20_+_52021000 | 1.09 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr5_-_65564733 | 1.08 |
ENSDART00000178162
|
CABZ01049025.1
|
ENSDARG00000107190 |
| chr20_+_26037296 | 1.08 |
ENSDART00000141265
|
znf106b
|
zinc finger protein 106b |
| chr13_+_28655433 | 1.08 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr15_-_5912917 | 1.07 |
|
|
|
| chr20_+_19094480 | 1.07 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr1_-_43011866 | 1.07 |
|
|
|
| chr2_-_58446028 | 1.06 |
ENSDART00000111651
|
ENSDARG00000078442
|
ENSDARG00000078442 |
| chr18_-_43890836 | 1.06 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_+_37648665 | 1.06 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr5_-_56641001 | 1.06 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr3_+_22204475 | 1.06 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr3_+_36830725 | 1.05 |
ENSDART00000150917
|
ENSDARG00000070174
|
ENSDARG00000070174 |
| chr2_-_38288904 | 1.04 |
|
|
|
| chr7_+_13238684 | 1.04 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr5_+_25408121 | 1.04 |
|
|
|
| chr14_-_26160351 | 1.03 |
|
|
|
| chr20_-_49067167 | 1.03 |
ENSDART00000163071
ENSDART00000170617 |
xrn2
|
5'-3' exoribonuclease 2 |
| chr9_-_7235272 | 1.03 |
ENSDART00000174720
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr7_+_45748086 | 1.03 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr7_-_24567002 | 1.03 |
ENSDART00000139455
|
fam113
|
family with sequence similarity 113 |
| chr3_+_54551887 | 1.02 |
ENSDART00000169663
|
CT573860.1
|
ENSDARG00000098327 |
| chr9_-_25517664 | 1.02 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr7_+_22524917 | 1.02 |
ENSDART00000112169
|
rbm4.2
|
RNA binding motif protein 4.2 |
| chr20_-_26148523 | 1.02 |
ENSDART00000141626
|
gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr8_+_7625791 | 1.02 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr14_+_33723918 | 1.02 |
ENSDART00000015670
|
med7
|
mediator complex subunit 7 |
| chr7_-_24604255 | 1.01 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr7_+_15197911 | 1.01 |
ENSDART00000046542
|
igf1rb
|
insulin-like growth factor 1b receptor |
| chr10_+_22566556 | 1.01 |
ENSDART00000079514
|
efnb3a
|
ephrin-B3a |
| chr1_-_51863300 | 1.00 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr10_+_19615343 | 1.00 |
|
|
|
| chr21_-_39501604 | 1.00 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr9_+_21548044 | 0.99 |
ENSDART00000147619
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr10_+_33629943 | 0.99 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr3_-_32457708 | 0.99 |
|
|
|
| chr25_+_7067043 | 0.99 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr21_+_599565 | 0.99 |
ENSDART00000177311
|
CABZ01072933.1
|
ENSDARG00000106379 |
| chr13_+_22567886 | 0.99 |
ENSDART00000137467
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr13_-_23626319 | 0.98 |
ENSDART00000124152
|
rgs17
|
regulator of G protein signaling 17 |
| chr6_+_8916643 | 0.98 |
|
|
|
| chr14_-_34293596 | 0.97 |
ENSDART00000128869
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr7_+_45748011 | 0.97 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr2_-_58805035 | 0.95 |
ENSDART00000159735
|
mau2
|
MAU2 sister chromatid cohesion factor |
| chr25_+_7067115 | 0.95 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr7_-_41446321 | 0.95 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.9 | 3.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.8 | 2.3 | GO:0071932 | replication fork reversal(GO:0071932) |
| 0.7 | 2.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.7 | 3.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.6 | 1.9 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.6 | 1.9 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.6 | 3.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 2.3 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 | 1.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.4 | 1.6 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.4 | 1.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.4 | 2.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 1.3 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 2.9 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 0.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.3 | 2.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 0.9 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.3 | 1.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 3.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 0.7 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 3.0 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 | 0.7 | GO:2000106 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.2 | 1.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.2 | 3.6 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 1.5 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.2 | 0.6 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 0.8 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 2.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 0.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.7 | GO:1990379 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 0.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 1.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.2 | 0.5 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.9 | GO:0051447 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of meiotic cell cycle(GO:0051447) |
| 0.2 | 1.7 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 1.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.2 | 1.6 | GO:0035283 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 1.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.6 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.6 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 2.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.0 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 0.7 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.0 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.1 | 1.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.7 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.4 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.8 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.5 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.4 | GO:1903335 | regulation of vacuolar transport(GO:1903335) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 4.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.4 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 0.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0032675 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 1.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.3 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.1 | GO:0090208 | positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.1 | 0.1 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 0.4 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.0 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.7 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.2 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.4 | GO:0046323 | type B pancreatic cell proliferation(GO:0044342) glucose import(GO:0046323) |
| 0.1 | 1.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:0097201 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 1.2 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 1.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.2 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.1 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.7 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.7 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) cellular response to dsRNA(GO:0071359) |
| 0.1 | 0.3 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.0 | GO:0072425 | G2 DNA damage checkpoint(GO:0031572) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 4.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.4 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.5 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 5.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 1.1 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 1.7 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 2.6 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 2.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.9 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 1.1 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.2 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.2 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 4.4 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.0 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0051170 | nuclear import(GO:0051170) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.7 | 4.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.6 | 2.3 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.4 | 2.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.4 | 3.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.4 | 1.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 1.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.3 | 2.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 2.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 3.6 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 2.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.9 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 2.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 5.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.4 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 1.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 8.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.6 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 3.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.5 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.8 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 4.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.3 | 3.9 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.8 | 2.3 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.7 | 2.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.7 | 2.1 | GO:0070336 | Y-form DNA binding(GO:0000403) double-strand/single-strand DNA junction binding(GO:0000406) flap-structured DNA binding(GO:0070336) |
| 0.6 | 3.6 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.5 | 2.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.5 | 5.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.5 | 1.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 1.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 1.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.4 | 1.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 3.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.4 | 2.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 1.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.3 | 0.9 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 3.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.3 | 0.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.3 | 0.8 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.3 | 1.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.9 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.9 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 7.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.6 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 0.6 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 1.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.9 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.6 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.2 | 0.7 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.2 | 0.5 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 4.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0045547 | polyprenyltransferase activity(GO:0002094) dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.8 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.6 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.7 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 6.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.2 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.0 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.3 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 2.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 3.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 4.7 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 6.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 3.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0004532 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 4.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 0.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 5.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.9 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.1 | 3.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 1.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |