Project
DANIO-CODE
Navigation
Downloads
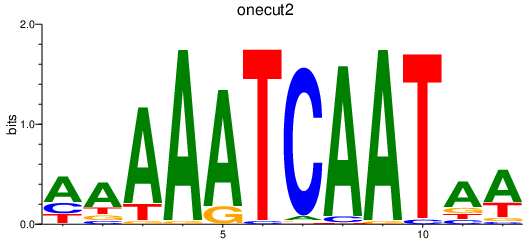
Results for onecut2
Z-value: 0.75
Transcription factors associated with onecut2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut2
|
ENSDARG00000090387 | one cut homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut2 | dr10_dc_chr21_-_1716495_1716567 | 0.36 | 1.7e-01 | Click! |
Activity profile of onecut2 motif
Sorted Z-values of onecut2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_14821305 | 2.34 |
|
|
|
| chr14_+_7626822 | 1.24 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr11_+_36933891 | 1.21 |
ENSDART00000170542
|
ENSDARG00000100406
|
ENSDARG00000100406 |
| chr6_-_10593395 | 1.15 |
ENSDART00000020261
|
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr19_-_35512932 | 1.12 |
|
|
|
| chr2_-_32521879 | 1.08 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr5_+_49103778 | 1.06 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr21_-_20305406 | 1.05 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr4_-_9172622 | 0.96 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr15_-_33688186 | 0.95 |
|
|
|
| chr3_+_23557320 | 0.95 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr20_+_52740555 | 0.91 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_-_33212897 | 0.89 |
ENSDART00000081170
|
cux1a
|
cut-like homeobox 1a |
| chr13_+_24271932 | 0.89 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr2_+_5651428 | 0.84 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr4_+_13811932 | 0.83 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr6_-_6895376 | 0.80 |
ENSDART00000081761
|
bin1b
|
bridging integrator 1b |
| chr4_-_17226968 | 0.78 |
ENSDART00000177956
ENSDART00000177406 |
BX547928.1
|
ENSDARG00000106175 |
| chr16_+_46526268 | 0.77 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr15_-_26619404 | 0.77 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr23_-_45250886 | 0.77 |
ENSDART00000163367
|
ENSDARG00000103727
|
ENSDARG00000103727 |
| chr16_+_22672315 | 0.76 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr5_+_23675413 | 0.73 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr11_-_11534819 | 0.73 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr7_-_25623974 | 0.71 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr21_+_19034242 | 0.70 |
ENSDART00000123309
|
nkx6.1
|
NK6 homeobox 1 |
| chr19_-_41887040 | 0.70 |
ENSDART00000062026
|
dlx5a
|
distal-less homeobox 5a |
| chr10_-_7827244 | 0.70 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr2_+_45447068 | 0.70 |
ENSDART00000083957
ENSDART00000160867 |
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr6_+_13833991 | 0.69 |
|
|
|
| chr4_-_10600228 | 0.66 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr11_+_42821961 | 0.66 |
ENSDART00000013642
|
foxg1b
|
forkhead box G1b |
| chr13_+_28286826 | 0.65 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr15_-_35394535 | 0.64 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr18_+_9762796 | 0.62 |
|
|
|
| chr10_-_33678206 | 0.62 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr23_-_17543731 | 0.59 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr7_-_57727148 | 0.58 |
|
|
|
| chr6_-_6895175 | 0.58 |
ENSDART00000149232
ENSDART00000150033 |
bin1b
|
bridging integrator 1b |
| chr17_+_32550616 | 0.56 |
|
|
|
| chr1_-_22466834 | 0.56 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr6_+_38882803 | 0.55 |
ENSDART00000132798
|
bin2b
|
bridging integrator 2b |
| chr5_-_45729685 | 0.54 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr3_-_28534446 | 0.54 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr19_+_19939410 | 0.54 |
|
|
|
| chr8_+_37136126 | 0.53 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr11_-_30364563 | 0.52 |
ENSDART00000165501
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr18_+_22367874 | 0.52 |
|
|
|
| chr25_+_18866796 | 0.51 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr21_-_27845076 | 0.51 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr5_-_62592627 | 0.50 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr4_-_9172550 | 0.49 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr1_+_50395721 | 0.48 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr11_+_30569525 | 0.48 |
|
|
|
| chr10_+_35479226 | 0.45 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr8_-_23060397 | 0.45 |
ENSDART00000135764
|
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr1_+_45267035 | 0.44 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr23_+_28843683 | 0.43 |
ENSDART00000132179
|
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr16_-_16793728 | 0.42 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr12_+_26376471 | 0.42 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr24_-_38068071 | 0.40 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr8_-_14446883 | 0.40 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr19_-_19282688 | 0.40 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr15_+_19523309 | 0.39 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr17_-_49931552 | 0.37 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr7_-_71192839 | 0.35 |
ENSDART00000124827
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr6_-_6895263 | 0.34 |
ENSDART00000149232
|
bin1b
|
bridging integrator 1b |
| chr18_-_25064913 | 0.33 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr14_-_2430754 | 0.33 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr20_-_6822668 | 0.32 |
ENSDART00000169569
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr11_+_19894502 | 0.31 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr14_-_32543646 | 0.30 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr7_+_25794014 | 0.29 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr6_+_46429927 | 0.29 |
ENSDART00000064865
ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr17_+_8018808 | 0.29 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr10_-_24422257 | 0.29 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr3_-_27989221 | 0.28 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr10_-_42157835 | 0.28 |
ENSDART00000141500
ENSDART00000156626 |
BX005309.1
|
ENSDARG00000093535 |
| chr3_-_22098046 | 0.28 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| KN149707v1_-_2099 | 0.27 |
|
|
|
| chr22_-_20141216 | 0.26 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr7_+_35243375 | 0.25 |
ENSDART00000160065
|
BX294178.1
|
ENSDARG00000101818 |
| chr3_-_6555471 | 0.24 |
ENSDART00000157771
ENSDART00000166758 |
sumo2b
|
small ubiquitin-like modifier 2b |
| chr4_-_2937706 | 0.23 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr14_+_21401757 | 0.23 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr21_+_3951876 | 0.21 |
ENSDART00000167791
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr4_-_10600179 | 0.21 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr1_+_45266889 | 0.20 |
ENSDART00000163725
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr2_-_39793399 | 0.20 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr6_-_2501966 | 0.19 |
ENSDART00000155109
|
hyi
|
hydroxypyruvate isomerase |
| chr16_+_43818521 | 0.18 |
ENSDART00000149154
|
si:dkey-122a22.2
|
si:dkey-122a22.2 |
| chr17_+_41034649 | 0.18 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr2_-_57133471 | 0.17 |
|
|
|
| chr5_+_19153421 | 0.17 |
|
|
|
| chr3_-_27989425 | 0.16 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_-_13256905 | 0.15 |
ENSDART00000026593
ENSDART00000150577 |
grip1
|
glutamate receptor interacting protein 1 |
| chr4_-_2937838 | 0.15 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr10_-_24422496 | 0.14 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr15_+_29241472 | 0.12 |
|
|
|
| chr12_-_34621359 | 0.12 |
|
|
|
| chr7_-_33080261 | 0.11 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr8_-_18579947 | 0.11 |
|
|
|
| chr18_-_17048300 | 0.10 |
ENSDART00000165479
|
has3
|
hyaluronan synthase 3 |
| chr1_-_20716842 | 0.10 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr6_+_12999420 | 0.10 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr4_-_2937524 | 0.09 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr10_-_15447374 | 0.08 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr18_-_44854305 | 0.07 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr21_+_30050280 | 0.07 |
|
|
|
| chr20_-_6822718 | 0.07 |
ENSDART00000169569
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr13_-_32517896 | 0.06 |
ENSDART00000132820
|
bend3
|
BEN domain containing 3 |
| chr1_-_53465275 | 0.06 |
|
|
|
| chr18_-_25065264 | 0.03 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr16_-_14506818 | 0.03 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr16_+_26147677 | 0.02 |
ENSDART00000113534
|
mrpl17
|
mitochondrial ribosomal protein L17 |
| chr13_-_15863571 | 0.00 |
ENSDART00000016430
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr5_-_23171454 | 0.00 |
ENSDART00000135153
|
TBC1D8B
|
TBC1 domain family member 8B |
| chr7_-_67741663 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.9 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.3 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.7 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.7 | GO:0050796 | pancreatic A cell differentiation(GO:0003310) regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 1.6 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.9 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.9 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0048899 | anterior lateral line development(GO:0048899) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.5 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |