Project
DANIO-CODE
Navigation
Downloads
Results for onecut3b
Z-value: 0.73
Transcription factors associated with onecut3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut3b
|
ENSDARG00000091059 | one cut homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut3b | dr10_dc_chr2_+_57369722_57369726 | 0.76 | 6.6e-04 | Click! |
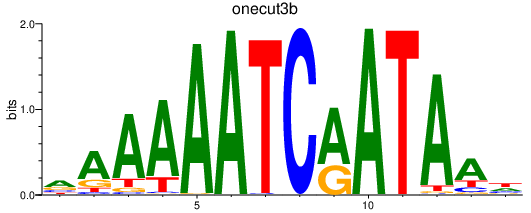
Activity profile of onecut3b motif
Sorted Z-values of onecut3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_23175007 | 2.44 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr10_+_10843384 | 2.14 |
ENSDART00000130283
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr24_-_23174888 | 1.85 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr10_-_33678206 | 1.75 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr15_-_26619404 | 1.72 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_14821305 | 1.65 |
|
|
|
| chr1_-_35419335 | 1.63 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr21_-_20305406 | 1.58 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr23_+_36079164 | 1.41 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr21_+_27346176 | 1.33 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr10_-_28949111 | 1.28 |
ENSDART00000030138
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr21_+_17731439 | 1.26 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr17_-_20877606 | 1.24 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr3_+_2013046 | 1.24 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr10_-_24422257 | 1.23 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr2_-_32521879 | 1.20 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr6_+_13833991 | 1.19 |
|
|
|
| chr11_+_6809190 | 1.17 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_+_55598181 | 1.17 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr19_+_19939410 | 1.16 |
|
|
|
| chr19_-_22182031 | 1.16 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr15_-_35034668 | 1.14 |
ENSDART00000153787
|
rnf183
|
ring finger protein 183 |
| chr16_+_33702010 | 1.13 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr16_+_19926776 | 1.08 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr11_+_36933891 | 1.07 |
ENSDART00000170542
|
ENSDARG00000100406
|
ENSDARG00000100406 |
| chr4_-_17226968 | 1.07 |
ENSDART00000177956
ENSDART00000177406 |
BX547928.1
|
ENSDARG00000106175 |
| chr13_+_24271932 | 1.06 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr5_+_18740715 | 1.04 |
ENSDART00000089173
|
atp8b5a
|
ATPase, class I, type 8B, member 5a |
| chr16_-_55234426 | 0.95 |
ENSDART00000078887
|
tmem222a
|
transmembrane protein 222a |
| chr2_+_5975726 | 0.92 |
ENSDART00000147831
|
ENSDARG00000056478
|
ENSDARG00000056478 |
| chr10_-_31838886 | 0.92 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_+_22672315 | 0.90 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr15_+_6070537 | 0.88 |
ENSDART00000123797
|
pcp4b
|
Purkinje cell protein 4b |
| chr10_-_7827244 | 0.88 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr2_+_45447068 | 0.85 |
ENSDART00000083957
ENSDART00000160867 |
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr3_+_25992836 | 0.85 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr18_+_5707066 | 0.85 |
|
|
|
| chr15_-_22978 | 0.84 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr19_+_31989527 | 0.82 |
ENSDART00000088401
|
acot13
|
acyl-CoA thioesterase 13 |
| chr11_+_42821961 | 0.81 |
ENSDART00000013642
|
foxg1b
|
forkhead box G1b |
| chr16_-_2412667 | 0.81 |
ENSDART00000103722
|
lyplal1
|
lysophospholipase-like 1 |
| chr7_-_57727148 | 0.81 |
|
|
|
| chr2_+_47727856 | 0.80 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr5_-_56981925 | 0.80 |
|
|
|
| chr3_-_28534446 | 0.78 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr5_-_70688185 | 0.76 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr19_+_26065401 | 0.73 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr20_-_25727145 | 0.73 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr25_+_35799816 | 0.72 |
ENSDART00000103016
|
zgc:173552
|
zgc:173552 |
| chr19_-_19282688 | 0.72 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_-_25695574 | 0.68 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr16_+_33701759 | 0.67 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr7_-_73631286 | 0.66 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr3_+_23561215 | 0.64 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr10_-_33212897 | 0.63 |
ENSDART00000081170
|
cux1a
|
cut-like homeobox 1a |
| chr4_+_13811932 | 0.62 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr23_-_36805789 | 0.62 |
|
|
|
| chr9_+_7745593 | 0.61 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr12_-_37921731 | 0.59 |
|
|
|
| chr18_+_22367874 | 0.59 |
|
|
|
| KN150672v1_-_47685 | 0.58 |
|
|
|
| chr3_-_30303173 | 0.58 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr20_-_23272102 | 0.58 |
ENSDART00000167197
|
spata18
|
spermatogenesis associated 18 |
| chr1_+_18639896 | 0.57 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr25_-_10992022 | 0.56 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr3_+_23561306 | 0.56 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr23_-_45250886 | 0.56 |
ENSDART00000163367
|
ENSDARG00000103727
|
ENSDARG00000103727 |
| chr8_-_14446883 | 0.54 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr8_-_23060397 | 0.52 |
ENSDART00000135764
|
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr22_-_16468179 | 0.51 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr6_-_2501966 | 0.51 |
ENSDART00000155109
|
hyi
|
hydroxypyruvate isomerase |
| chr25_+_21919595 | 0.51 |
ENSDART00000156517
|
ENSDARG00000062199
|
ENSDARG00000062199 |
| chr12_-_34621359 | 0.50 |
|
|
|
| chr20_+_52740555 | 0.50 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr14_-_32543646 | 0.49 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr13_+_49436597 | 0.48 |
ENSDART00000037559
ENSDART00000168799 ENSDART00000097845 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr3_-_22098046 | 0.48 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr3_+_32557992 | 0.45 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr10_+_35479226 | 0.45 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr4_+_8007802 | 0.45 |
ENSDART00000014036
|
optn
|
optineurin |
| chr23_+_28843683 | 0.45 |
ENSDART00000132179
|
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr4_-_998370 | 0.44 |
|
|
|
| chr23_+_34037058 | 0.43 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr1_-_22466834 | 0.42 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr10_-_28949067 | 0.42 |
ENSDART00000030138
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr1_-_35419273 | 0.42 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr6_-_17699766 | 0.40 |
ENSDART00000170597
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr11_-_30364563 | 0.39 |
ENSDART00000165501
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr11_-_10814101 | 0.39 |
ENSDART00000091923
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr18_+_3482740 | 0.39 |
ENSDART00000158763
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr2_+_13779498 | 0.38 |
ENSDART00000149309
|
cfap57
|
cilia and flagella associated protein 57 |
| chr20_-_31028201 | 0.38 |
ENSDART00000146376
|
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr13_-_9563572 | 0.37 |
ENSDART00000142629
|
ENSDARG00000092071
|
ENSDARG00000092071 |
| chr1_+_23093475 | 0.36 |
|
|
|
| chr3_-_15959984 | 0.36 |
ENSDART00000140376
|
lasp1
|
LIM and SH3 protein 1 |
| chr16_+_24045774 | 0.35 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr8_-_14014576 | 0.35 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr11_+_24826881 | 0.33 |
ENSDART00000157435
|
ndrg3a
|
ndrg family member 3a |
| chr4_-_74907755 | 0.33 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr16_+_4825665 | 0.32 |
|
|
|
| chr2_-_39793399 | 0.31 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr3_+_42212805 | 0.30 |
ENSDART00000169820
|
psmg3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr21_+_3951876 | 0.27 |
ENSDART00000167791
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr21_+_1504942 | 0.27 |
ENSDART00000130274
|
rab27b
|
RAB27B, member RAS oncogene family |
| chr21_+_6107190 | 0.26 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr12_-_13298628 | 0.25 |
ENSDART00000105903
|
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_+_35243375 | 0.25 |
ENSDART00000160065
|
BX294178.1
|
ENSDARG00000101818 |
| chr11_+_17805723 | 0.25 |
|
|
|
| chr6_+_38882803 | 0.23 |
ENSDART00000132798
|
bin2b
|
bridging integrator 2b |
| chr9_-_264078 | 0.23 |
ENSDART00000166231
|
pcbp2
|
poly(rC) binding protein 2 |
| chr18_-_19495618 | 0.22 |
ENSDART00000177690
|
lctlb
|
lactase-like b |
| chr25_-_34541162 | 0.22 |
ENSDART00000125755
|
hist1h4l
|
histone 1, H4, like |
| chr3_-_27989221 | 0.22 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_-_26912259 | 0.21 |
ENSDART00000147735
|
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr25_-_36499819 | 0.20 |
ENSDART00000150178
|
wwox
|
WW domain containing oxidoreductase |
| chr19_-_12106837 | 0.20 |
|
|
|
| chr18_-_20570874 | 0.18 |
ENSDART00000141367
|
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr13_-_31036047 | 0.18 |
ENSDART00000134640
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr12_-_37921503 | 0.18 |
|
|
|
| chr5_-_68439806 | 0.17 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr10_-_24422496 | 0.16 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr1_-_20716842 | 0.15 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr19_-_46427187 | 0.15 |
ENSDART00000169957
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr3_+_39816596 | 0.13 |
ENSDART00000099754
|
ccdc97
|
coiled-coil domain containing 97 |
| chr21_+_1504883 | 0.13 |
ENSDART00000130274
|
rab27b
|
RAB27B, member RAS oncogene family |
| chr18_-_44854305 | 0.11 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr21_+_6107404 | 0.11 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr20_+_19093073 | 0.11 |
ENSDART00000147105
|
tdh
|
L-threonine dehydrogenase |
| chr23_-_28099391 | 0.10 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr20_-_2991247 | 0.09 |
ENSDART00000160701
|
ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr15_-_33886897 | 0.09 |
|
|
|
| chr15_-_7622212 | 0.08 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr20_-_43182331 | 0.08 |
ENSDART00000172655
|
snx9b
|
sorting nexin 9b |
| chr23_-_28099284 | 0.08 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr24_+_33478309 | 0.07 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr12_+_48888044 | 0.06 |
ENSDART00000109315
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr6_-_16328987 | 0.04 |
ENSDART00000083305
|
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr20_-_25727347 | 0.04 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr15_+_19523309 | 0.04 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr9_-_44487864 | 0.03 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr20_+_25727285 | 0.02 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr20_+_34248780 | 0.02 |
ENSDART00000152870
|
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr7_+_13130332 | 0.01 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr3_-_27989425 | 0.01 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr24_+_9740818 | 0.00 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 2.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 1.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 0.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 0.5 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.6 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 1.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.9 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.1 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.6 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.8 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.0 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 1.1 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.8 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 3.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |