Project
DANIO-CODE
Navigation
Downloads
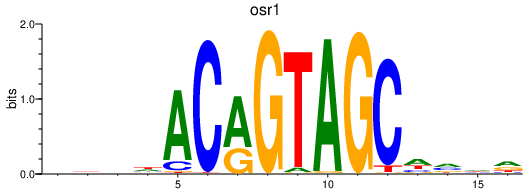
Results for osr1
Z-value: 0.46
Transcription factors associated with osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr1
|
ENSDARG00000014091 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr1 | dr10_dc_chr13_+_32013916_32014006 | -0.34 | 2.0e-01 | Click! |
Activity profile of osr1 motif
Sorted Z-values of osr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of osr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_33418562 | 0.85 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr25_-_34638406 | 0.65 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr18_-_45750 | 0.64 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_29768364 | 0.49 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr6_-_40886902 | 0.43 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr25_-_7490206 | 0.42 |
ENSDART00000163018
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr19_+_24455545 | 0.42 |
ENSDART00000144541
|
rit1
|
Ras-like without CAAX 1 |
| chr18_-_1084561 | 0.39 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr4_-_1975473 | 0.36 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr24_-_9819862 | 0.36 |
ENSDART00000092975
|
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr6_-_43297150 | 0.35 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr16_+_51696466 | 0.33 |
|
|
|
| chr2_+_56339323 | 0.32 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr10_-_13281047 | 0.30 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr16_+_46718177 | 0.30 |
ENSDART00000058325
|
lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr11_+_26366532 | 0.30 |
ENSDART00000159505
|
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr10_+_4964156 | 0.28 |
ENSDART00000109882
|
lyrm7
|
LYR motif containing 7 |
| chr7_-_3332675 | 0.28 |
|
|
|
| chr9_-_34459799 | 0.27 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr19_+_31989527 | 0.27 |
ENSDART00000088401
|
acot13
|
acyl-CoA thioesterase 13 |
| chr21_-_25258473 | 0.27 |
ENSDART00000101153
ENSDART00000147860 ENSDART00000087910 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr19_-_1376498 | 0.26 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr23_-_5188577 | 0.25 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr14_+_4044357 | 0.25 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr3_-_54884533 | 0.24 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr13_-_30741976 | 0.23 |
ENSDART00000142535
|
CR762493.1
|
ENSDARG00000092411 |
| chr3_-_14994229 | 0.23 |
ENSDART00000161213
|
fam173a
|
family with sequence similarity 173, member A |
| chr11_-_36017410 | 0.22 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr24_+_9553580 | 0.22 |
ENSDART00000082411
|
topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr20_-_54645287 | 0.22 |
ENSDART00000153389
|
yy1b
|
YY1 transcription factor b |
| chr3_+_25776714 | 0.22 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr14_-_46649324 | 0.22 |
|
|
|
| chr18_+_14677042 | 0.21 |
ENSDART00000138995
|
vps9d1
|
VPS9 domain containing 1 |
| chr17_-_26519469 | 0.21 |
ENSDART00000155692
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr6_-_49160398 | 0.21 |
ENSDART00000143241
|
tspan2a
|
tetraspanin 2a |
| chr19_-_6215704 | 0.21 |
ENSDART00000104978
|
cica
|
capicua transcriptional repressor a |
| chr3_-_57694604 | 0.21 |
ENSDART00000013376
|
cant1a
|
calcium activated nucleotidase 1a |
| KN150683v1_+_13359 | 0.21 |
|
|
|
| chr10_+_19597171 | 0.20 |
ENSDART00000063810
ENSDART00000063806 |
atp6v1b2
|
ATPase, H+ transporting, lysosomal, V1 subunit B2 |
| chr12_-_45714172 | 0.20 |
ENSDART00000149044
|
pax2b
|
paired box 2b |
| chr25_+_20021806 | 0.20 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr10_+_29251398 | 0.19 |
ENSDART00000025227
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr23_+_4385722 | 0.19 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr17_-_16079935 | 0.19 |
|
|
|
| chr11_-_36017515 | 0.18 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr19_+_43745515 | 0.18 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr16_+_53602834 | 0.18 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr23_+_20586769 | 0.18 |
ENSDART00000079591
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr12_+_21176669 | 0.18 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr16_+_35391136 | 0.17 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr2_-_37479961 | 0.17 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr22_-_11596329 | 0.17 |
ENSDART00000063133
|
gcga
|
glucagon a |
| chr22_-_21289850 | 0.17 |
ENSDART00000135388
|
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr14_-_26238386 | 0.17 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr21_-_7334721 | 0.17 |
ENSDART00000136671
|
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr1_-_57329034 | 0.16 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr15_+_20416518 | 0.16 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr13_+_25266451 | 0.16 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr3_-_26052601 | 0.16 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_23650217 | 0.16 |
ENSDART00000080609
|
mrps31
|
mitochondrial ribosomal protein S31 |
| KN150339v1_-_47655 | 0.16 |
|
|
|
| chr2_-_14833381 | 0.15 |
|
|
|
| chr5_-_55843553 | 0.15 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr14_+_29428950 | 0.15 |
ENSDART00000105898
|
ENSDARG00000071567
|
ENSDARG00000071567 |
| chr18_-_46260401 | 0.15 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr3_-_30494171 | 0.15 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr7_-_34656337 | 0.14 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3a |
| chr22_-_19077300 | 0.14 |
ENSDART00000166295
|
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr18_+_45680282 | 0.14 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr3_-_11409342 | 0.14 |
|
|
|
| chr11_-_40192951 | 0.14 |
ENSDART00000102750
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr3_+_56008829 | 0.14 |
|
|
|
| chr4_-_74907755 | 0.14 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr9_-_6524104 | 0.14 |
ENSDART00000158468
|
nck2a
|
NCK adaptor protein 2a |
| chr3_-_29992106 | 0.14 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr21_-_33998148 | 0.14 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr21_+_21158761 | 0.14 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr18_+_21419735 | 0.14 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_-_29992171 | 0.14 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr5_-_56277572 | 0.14 |
|
|
|
| chr9_-_18561215 | 0.14 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr24_+_10276926 | 0.13 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr16_+_29574449 | 0.13 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr13_+_36797201 | 0.13 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr25_-_13562693 | 0.13 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr6_+_54568388 | 0.13 |
ENSDART00000093199
|
tead3b
|
TEA domain family member 3 b |
| chr7_-_28771601 | 0.13 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr14_+_14541725 | 0.13 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr7_+_41871444 | 0.13 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr9_-_21287478 | 0.13 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr7_+_24119940 | 0.13 |
ENSDART00000108753
|
poln
|
polymerase (DNA directed) nu |
| chr6_-_11576632 | 0.13 |
ENSDART00000151717
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr16_-_22947908 | 0.13 |
ENSDART00000133910
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_+_13039609 | 0.13 |
ENSDART00000109876
ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr2_-_22707734 | 0.13 |
ENSDART00000158486
|
sep15
|
selenoprotein 15 |
| chr18_-_1084697 | 0.13 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr12_-_45897929 | 0.13 |
|
|
|
| chr22_-_9809500 | 0.12 |
|
|
|
| chr22_+_22393190 | 0.12 |
ENSDART00000105600
|
ddx59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
| chr7_-_41871376 | 0.12 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| KN150456v1_-_16664 | 0.12 |
|
|
|
| chr11_-_1409262 | 0.12 |
ENSDART00000155269
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr1_+_50763649 | 0.12 |
ENSDART00000074294
|
actr2a
|
ARP2 actin related protein 2a homolog |
| chr12_-_1544844 | 0.11 |
|
|
|
| chr7_-_54990861 | 0.11 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr2_-_17721575 | 0.11 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr7_+_24458357 | 0.11 |
ENSDART00000111542
|
shroom4
|
shroom family member 4 |
| chr13_-_14175196 | 0.11 |
|
|
|
| KN150703v1_-_47381 | 0.11 |
ENSDART00000168391
|
CABZ01081752.1
|
ENSDARG00000099245 |
| chr5_+_29231012 | 0.11 |
ENSDART00000159327
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr4_+_10618294 | 0.11 |
ENSDART00000067251
|
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr21_-_32003326 | 0.10 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr10_+_2559774 | 0.10 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr18_-_15361133 | 0.10 |
ENSDART00000019818
|
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr13_-_42453219 | 0.10 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| KN149895v1_+_26398 | 0.10 |
|
|
|
| chr21_+_13689239 | 0.10 |
ENSDART00000112451
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr8_+_3391493 | 0.10 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr8_+_54058732 | 0.10 |
|
|
|
| chr2_-_41873625 | 0.10 |
ENSDART00000155577
|
ENSDARG00000056490
|
ENSDARG00000056490 |
| chr16_-_43090120 | 0.10 |
ENSDART00000155637
|
BX005234.1
|
ENSDARG00000097761 |
| chr7_-_33558939 | 0.10 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr12_-_4800362 | 0.10 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr15_-_4424628 | 0.10 |
ENSDART00000062874
|
atp1b3b
|
ATPase, Na+/K+ transporting, beta 3b polypeptide |
| chr18_+_30868972 | 0.10 |
ENSDART00000176162
|
foxf1
|
forkhead box F1 |
| chr2_+_50892294 | 0.10 |
ENSDART00000018150
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr5_-_66070385 | 0.09 |
ENSDART00000032909
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr3_+_17594555 | 0.09 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr14_-_46649231 | 0.09 |
|
|
|
| chr25_-_21692269 | 0.09 |
|
|
|
| chr24_+_10273081 | 0.09 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr5_-_54578261 | 0.09 |
ENSDART00000016995
ENSDART00000134937 |
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr15_+_20416637 | 0.09 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr23_-_29643223 | 0.09 |
ENSDART00000133576
|
nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr14_-_46649445 | 0.09 |
|
|
|
| chr5_+_59845172 | 0.09 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr15_-_19192862 | 0.09 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr16_+_35391089 | 0.09 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr2_+_10362661 | 0.09 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr18_+_30529524 | 0.08 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr15_-_19192933 | 0.08 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr19_+_47707575 | 0.08 |
ENSDART00000163591
ENSDART00000159860 ENSDART00000138909 ENSDART00000158588 ENSDART00000170847 |
tpmt.2
|
thiopurine S-methyltransferase, tandem duplicate 2 |
| chr7_+_23388546 | 0.08 |
|
|
|
| chr25_+_21001071 | 0.08 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr5_-_6054581 | 0.08 |
|
|
|
| KN150289v1_-_7882 | 0.08 |
|
|
|
| chr25_+_21001126 | 0.08 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr14_+_35560934 | 0.08 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr16_-_6318609 | 0.08 |
|
|
|
| chr1_+_25911314 | 0.07 |
ENSDART00000011645
|
coro2a
|
coronin, actin binding protein, 2A |
| chr8_-_2447758 | 0.07 |
ENSDART00000101125
|
rpl6
|
ribosomal protein L6 |
| chr1_+_9202797 | 0.07 |
ENSDART00000135702
|
rgs11
|
regulator of G protein signaling 11 |
| chr2_-_9848848 | 0.07 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr22_-_38964356 | 0.07 |
ENSDART00000131462
|
rbbp9
|
retinoblastoma binding protein 9 |
| chr7_+_33044157 | 0.07 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr25_+_17764396 | 0.07 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr16_+_25425398 | 0.07 |
|
|
|
| chr11_-_36503530 | 0.07 |
ENSDART00000002055
|
cacna2d3
|
calcium channel, voltage dependent, alpha2/delta subunit 3 |
| chr23_-_20412221 | 0.07 |
ENSDART00000132769
|
CR749762.2
|
ENSDARG00000093223 |
| chr15_+_29126271 | 0.06 |
ENSDART00000144546
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr6_+_44891187 | 0.06 |
|
|
|
| chr15_+_20961370 | 0.06 |
ENSDART00000154854
|
CR848784.5
|
ENSDARG00000097996 |
| chr9_+_33310981 | 0.06 |
|
|
|
| chr15_-_9616877 | 0.06 |
ENSDART00000168668
|
gab2
|
GRB2-associated binding protein 2 |
| chr21_-_33998091 | 0.06 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr7_+_38044801 | 0.06 |
ENSDART00000160197
|
BX323855.1
|
ENSDARG00000098940 |
| chr7_-_33559032 | 0.06 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr5_-_19342462 | 0.06 |
ENSDART00000159309
|
trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr18_+_35994461 | 0.06 |
|
|
|
| chr15_+_20416591 | 0.06 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr8_+_2471513 | 0.06 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr4_-_6558919 | 0.05 |
ENSDART00000142087
|
foxp2
|
forkhead box P2 |
| chr3_-_26052785 | 0.05 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_-_22875246 | 0.05 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr16_+_21065200 | 0.05 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr10_+_15078155 | 0.05 |
|
|
|
| chr5_-_36348618 | 0.05 |
|
|
|
| chr2_-_51362397 | 0.05 |
ENSDART00000167172
|
THEM6
|
thioesterase superfamily member 6 |
| chr22_-_4622401 | 0.05 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr8_-_43842142 | 0.05 |
ENSDART00000134073
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr13_+_24249677 | 0.05 |
ENSDART00000022682
|
pdcd2
|
programmed cell death 2 |
| chr24_+_35173816 | 0.05 |
|
|
|
| chr23_-_35904414 | 0.05 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr21_-_11564398 | 0.05 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr13_+_40560230 | 0.05 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr11_-_33697442 | 0.05 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr25_+_21001016 | 0.05 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr15_+_46799979 | 0.04 |
|
|
|
| chr3_-_57691670 | 0.04 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr21_-_11564441 | 0.04 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr10_-_7743194 | 0.04 |
ENSDART00000018017
|
gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr2_-_25475213 | 0.04 |
|
|
|
| chr21_-_28700622 | 0.04 |
ENSDART00000098696
|
nrg2a
|
neuregulin 2a |
| chr2_-_14833347 | 0.04 |
|
|
|
| chr19_-_9584480 | 0.04 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr5_-_39598763 | 0.04 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr14_-_40022693 | 0.04 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr1_-_18084246 | 0.04 |
|
|
|
| chr3_+_32440145 | 0.04 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr8_+_7655655 | 0.04 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 0.2 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0097704 | response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0070166 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |