Project
DANIO-CODE
Navigation
Downloads
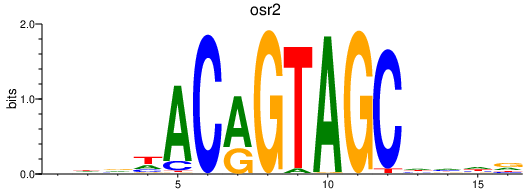
Results for osr2
Z-value: 0.46
Transcription factors associated with osr2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr2
|
ENSDARG00000038006 | odd-skipped related transciption factor 2 |
Activity profile of osr2 motif
Sorted Z-values of osr2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of osr2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_45750 | 1.38 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr7_-_33558939 | 1.14 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr14_+_35906366 | 0.78 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr22_-_11596329 | 0.76 |
ENSDART00000063133
|
gcga
|
glucagon a |
| chr10_-_13281047 | 0.68 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr18_-_1084561 | 0.67 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr10_+_38568464 | 0.64 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr23_+_35996491 | 0.61 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr16_+_35391136 | 0.55 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr6_-_40886902 | 0.55 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr14_+_8198019 | 0.50 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr7_-_33559032 | 0.48 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr8_+_3391493 | 0.47 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr18_-_1084697 | 0.46 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr22_-_4622401 | 0.40 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr16_+_35391089 | 0.39 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr2_+_4144790 | 0.36 |
ENSDART00000160806
|
mkxb
|
mohawk homeobox b |
| chr16_+_51696466 | 0.35 |
|
|
|
| chr5_-_54383466 | 0.34 |
|
|
|
| chr17_-_16079935 | 0.34 |
|
|
|
| chr3_-_11409342 | 0.32 |
|
|
|
| chr11_+_31022048 | 0.30 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr11_-_36017515 | 0.29 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr18_-_45892 | 0.29 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_+_21065200 | 0.29 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr3_-_27989221 | 0.24 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr23_-_35904414 | 0.22 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr19_+_47707575 | 0.17 |
ENSDART00000163591
ENSDART00000159860 ENSDART00000138909 ENSDART00000158588 ENSDART00000170847 |
tpmt.2
|
thiopurine S-methyltransferase, tandem duplicate 2 |
| chr10_-_41742329 | 0.15 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr11_-_36017410 | 0.13 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr3_-_27989425 | 0.10 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_18410629 | 0.09 |
|
|
|
| chr7_-_41871376 | 0.08 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr19_-_6215704 | 0.07 |
ENSDART00000104978
|
cica
|
capicua transcriptional repressor a |
| chr6_-_43297150 | 0.06 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr14_+_23419894 | 0.05 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr5_+_6054781 | 0.04 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr3_-_27989329 | 0.00 |
ENSDART00000151143
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 1.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.6 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.5 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.8 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.8 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |