Project
DANIO-CODE
Navigation
Downloads
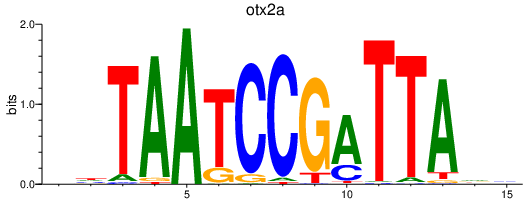
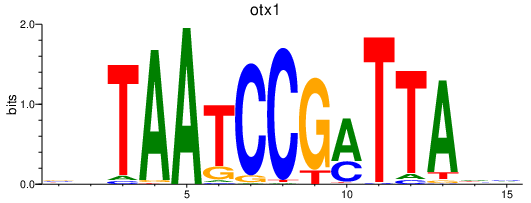
Results for otx2a_otx1
Z-value: 0.53
Transcription factors associated with otx2a_otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx2a
|
ENSDARG00000030703 | orthodenticle homeobox 2a |
|
otx1
|
ENSDARG00000094992 | orthodenticle homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx1a | dr10_dc_chr1_+_50277442_50277494 | 0.64 | 7.9e-03 | Click! |
| otx1b | dr10_dc_chr17_+_24300451_24300487 | -0.51 | 4.4e-02 | Click! |
Activity profile of otx2a_otx1 motif
Sorted Z-values of otx2a_otx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of otx2a_otx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_34367105 | 1.92 |
ENSDART00000157625
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr25_+_18866796 | 1.14 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr19_+_2013850 | 1.10 |
ENSDART00000048850
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr1_-_674449 | 0.99 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr5_-_64348528 | 0.89 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr9_-_30437160 | 0.85 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr3_+_34010958 | 0.84 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr5_+_45822196 | 0.79 |
|
|
|
| chr1_-_44888989 | 0.73 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr24_+_9335428 | 0.70 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr21_-_44610563 | 0.67 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr20_+_1251702 | 0.64 |
ENSDART00000064447
|
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr17_-_8111181 | 0.64 |
ENSDART00000149873
ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr7_+_38123920 | 0.63 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr24_-_8590572 | 0.63 |
ENSDART00000082349
|
tfap2a
|
transcription factor AP-2 alpha |
| chr8_+_31426328 | 0.61 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr2_-_23431625 | 0.57 |
ENSDART00000157318
|
BX548024.1
|
ENSDARG00000097593 |
| chr24_+_9335316 | 0.53 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr4_-_18466436 | 0.53 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr9_-_38232999 | 0.53 |
ENSDART00000133998
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr17_+_10437343 | 0.50 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr22_-_32553488 | 0.48 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr15_+_926222 | 0.45 |
ENSDART00000156395
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr2_-_49114158 | 0.44 |
|
|
|
| chr7_+_19857748 | 0.44 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr16_+_46530388 | 0.44 |
ENSDART00000058324
|
rpz4
|
rapunzel 4 |
| chr19_+_47846675 | 0.43 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr6_+_16279737 | 0.42 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr5_-_40451061 | 0.42 |
ENSDART00000083515
|
pdzd2
|
PDZ domain containing 2 |
| chr7_+_19857885 | 0.42 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr15_+_676493 | 0.42 |
|
|
|
| chr6_+_37677155 | 0.41 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr7_+_19858190 | 0.41 |
ENSDART00000179395
ENSDART00000113793 ENSDART00000169603 |
zgc:114045
|
zgc:114045 |
| chr23_-_3816492 | 0.41 |
ENSDART00000136394
|
hmga1a
|
high mobility group AT-hook 1a |
| chr5_-_56691295 | 0.40 |
ENSDART00000112793
|
cdc26
|
cell division cycle 26 homolog |
| chr4_-_29059042 | 0.40 |
ENSDART00000169971
|
CR387997.1
|
ENSDARG00000099486 |
| chr23_+_41129663 | 0.38 |
|
|
|
| chr7_-_35161302 | 0.38 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr19_-_7621797 | 0.36 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr6_-_53426323 | 0.36 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr7_-_46510632 | 0.35 |
|
|
|
| chr19_+_34582100 | 0.34 |
ENSDART00000135592
|
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr8_+_47909655 | 0.33 |
|
|
|
| chr14_-_9274939 | 0.33 |
|
|
|
| chr18_+_45680493 | 0.33 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr7_-_35136671 | 0.33 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr19_+_7234029 | 0.32 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr14_-_4166292 | 0.32 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr8_-_49754713 | 0.32 |
ENSDART00000074840
|
zgc:136564
|
zgc:136564 |
| chr24_+_20391758 | 0.31 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr14_-_25688444 | 0.31 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr10_+_39291358 | 0.31 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr21_-_36338704 | 0.31 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr9_+_1968455 | 0.31 |
|
|
|
| chr13_-_33269297 | 0.30 |
|
|
|
| chr5_-_40310798 | 0.29 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr5_-_32282404 | 0.29 |
ENSDART00000166698
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr9_-_2938937 | 0.28 |
|
|
|
| chr18_-_25002972 | 0.28 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr7_-_35136817 | 0.27 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr10_+_32160464 | 0.27 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr11_-_27455242 | 0.27 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr20_-_35109185 | 0.27 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr2_+_49683715 | 0.27 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr13_+_30097568 | 0.27 |
ENSDART00000134691
|
rps24
|
ribosomal protein S24 |
| chr4_-_20764499 | 0.27 |
ENSDART00000023337
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr22_-_31722970 | 0.27 |
ENSDART00000128247
|
lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr21_-_2116499 | 0.25 |
ENSDART00000167307
|
AL627305.3
|
ENSDARG00000102019 |
| chr17_-_9869142 | 0.25 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr4_+_67836787 | 0.25 |
ENSDART00000168400
|
znf1117
|
zinc finger protein 1117 |
| KN149874v1_-_3544 | 0.25 |
ENSDART00000165981
|
CABZ01046348.1
|
ENSDARG00000099481 |
| chr18_+_9762796 | 0.24 |
|
|
|
| chr21_-_25176853 | 0.24 |
ENSDART00000122513
|
rfc2
|
replication factor C (activator 1) 2 |
| chr4_+_28452948 | 0.23 |
ENSDART00000150879
|
znf1067
|
zinc finger protein 1067 |
| chr17_+_14878846 | 0.23 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr3_+_36282205 | 0.23 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr7_-_39107780 | 0.22 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr21_+_43706830 | 0.22 |
ENSDART00000126190
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr7_-_16346150 | 0.21 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr4_+_11054808 | 0.21 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr5_-_40310638 | 0.21 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_41791642 | 0.21 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr3_-_39287733 | 0.21 |
|
|
|
| chr19_-_20203902 | 0.19 |
ENSDART00000172668
|
CR382300.3
|
ENSDARG00000101330 |
| chr23_-_3816422 | 0.19 |
ENSDART00000132205
|
hmga1a
|
high mobility group AT-hook 1a |
| chr21_-_20728623 | 0.19 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr3_-_15849644 | 0.19 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_-_8961733 | 0.19 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| KN150529v1_+_16214 | 0.18 |
ENSDART00000177459
|
CABZ01084538.1
|
ENSDARG00000108298 |
| chr7_-_46510731 | 0.18 |
|
|
|
| chr18_+_23891068 | 0.18 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr8_+_25594106 | 0.18 |
|
|
|
| chr11_-_27455348 | 0.17 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr3_-_15850290 | 0.17 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr22_-_16128314 | 0.16 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_26246985 | 0.16 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr11_-_18073742 | 0.16 |
|
|
|
| chr9_-_46475828 | 0.16 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr17_+_50622804 | 0.16 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr1_-_37451594 | 0.16 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr4_-_29003925 | 0.16 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr4_+_17364740 | 0.16 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr10_+_26691086 | 0.15 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr3_+_32360862 | 0.15 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr8_-_45422915 | 0.15 |
ENSDART00000150067
|
ywhabb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b |
| chr3_-_12752075 | 0.15 |
ENSDART00000160522
|
ccdc137
|
coiled-coil domain containing 137 |
| chr16_-_48458505 | 0.15 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr1_+_35217331 | 0.15 |
|
|
|
| chr16_-_6450531 | 0.14 |
ENSDART00000060550
ENSDART00000145663 |
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr7_-_35136743 | 0.14 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr16_+_38209664 | 0.14 |
ENSDART00000160332
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr18_+_284567 | 0.14 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr3_+_1440282 | 0.13 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr18_-_10744619 | 0.13 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr9_-_1457681 | 0.13 |
ENSDART00000124777
ENSDART00000093412 |
osbpl6
|
oxysterol binding protein-like 6 |
| chr20_-_42481675 | 0.13 |
ENSDART00000015801
|
dcbld1
|
discoidin, CUB and LCCL domain containing 1 |
| chr2_+_58658615 | 0.13 |
|
|
|
| chr23_-_36342779 | 0.12 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr5_-_40310702 | 0.12 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr10_-_14530340 | 0.12 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr2_+_55859099 | 0.12 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr12_-_9046799 | 0.12 |
ENSDART00000125230
|
exoc6
|
exocyst complex component 6 |
| chr25_-_10533634 | 0.11 |
ENSDART00000153639
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr4_-_29488075 | 0.11 |
ENSDART00000167636
|
CR847895.4
|
ENSDARG00000101492 |
| chr19_-_26093745 | 0.11 |
ENSDART00000022676
|
asns
|
asparagine synthetase |
| chr18_+_23891152 | 0.10 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr11_+_18710724 | 0.10 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr25_-_14494641 | 0.10 |
|
|
|
| chr19_-_5141645 | 0.10 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr19_+_2803373 | 0.10 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr18_+_10816190 | 0.09 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr8_-_19872014 | 0.09 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr17_+_1950129 | 0.09 |
ENSDART00000110529
|
bub1bb
|
BUB1 mitotic checkpoint serine/threonine kinase Bb |
| chr8_+_53241686 | 0.08 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr20_-_29028920 | 0.08 |
ENSDART00000153082
|
susd6
|
sushi domain containing 6 |
| chr23_-_45686191 | 0.08 |
|
|
|
| chr4_+_36180573 | 0.08 |
ENSDART00000161159
|
ENSDARG00000102102
|
ENSDARG00000102102 |
| chr23_+_21565625 | 0.07 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr7_-_26246798 | 0.07 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr1_+_30941618 | 0.07 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr18_+_10815994 | 0.07 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr3_+_15605866 | 0.07 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr17_+_50622887 | 0.07 |
ENSDART00000134126
|
fermt2
|
fermitin family member 2 |
| chr25_-_10532989 | 0.07 |
ENSDART00000177834
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr15_+_20428823 | 0.07 |
ENSDART00000062676
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr21_-_44109455 | 0.07 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr19_+_34581867 | 0.07 |
ENSDART00000135592
|
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr15_-_26998580 | 0.06 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr10_-_14530112 | 0.06 |
ENSDART00000176126
|
galt
|
galactose-1-phosphate uridylyltransferase |
| chr8_-_29842274 | 0.06 |
ENSDART00000086044
ENSDART00000167487 |
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr9_+_21340251 | 0.06 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr21_+_36896321 | 0.06 |
ENSDART00000086001
|
arl10
|
ADP-ribosylation factor-like 10 |
| chr5_-_43160243 | 0.05 |
|
|
|
| chr2_+_20946939 | 0.05 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr5_+_32282547 | 0.05 |
ENSDART00000135459
|
rpl12
|
ribosomal protein L12 |
| chr11_-_12686320 | 0.05 |
ENSDART00000081319
|
zrsr2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr4_+_56350528 | 0.05 |
ENSDART00000167976
|
BX927290.1
|
ENSDARG00000098469 |
| chr1_-_17898961 | 0.05 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr5_+_61136704 | 0.05 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr10_+_39555088 | 0.04 |
ENSDART00000135756
|
kirrel3a
|
kin of IRRE like 3 a |
| chr7_+_19779221 | 0.04 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr2_+_58658858 | 0.04 |
|
|
|
| chr25_+_6607551 | 0.04 |
ENSDART00000176596
|
FQ377991.1
|
ENSDARG00000108452 |
| chr3_-_15060501 | 0.04 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr19_-_2222108 | 0.04 |
ENSDART00000127578
|
twistnb
|
TWIST neighbor |
| chr3_+_29849796 | 0.03 |
ENSDART00000151509
|
knca7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr24_-_21750082 | 0.03 |
|
|
|
| chr7_-_39107722 | 0.03 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr17_-_31041977 | 0.03 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr1_+_51998547 | 0.03 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr17_+_30431520 | 0.03 |
ENSDART00000153826
|
lpin1
|
lipin 1 |
| chr13_-_35924447 | 0.03 |
ENSDART00000134955
|
lgmn
|
legumain |
| chr5_-_68408171 | 0.02 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr10_+_3052919 | 0.02 |
ENSDART00000028870
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr11_+_6298048 | 0.02 |
ENSDART00000104409
ENSDART00000139882 |
ranbp3a
|
RAN binding protein 3a |
| chr9_+_3547661 | 0.02 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr5_+_34863704 | 0.02 |
ENSDART00000131286
|
erlin2
|
ER lipid raft associated 2 |
| chr6_-_7937201 | 0.02 |
|
|
|
| chr1_-_37451291 | 0.02 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr5_-_25133456 | 0.01 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr19_+_44288538 | 0.01 |
ENSDART00000139684
|
lypla2
|
lysophospholipase II |
| chr20_-_34868582 | 0.01 |
|
|
|
| chr18_+_23890892 | 0.01 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr1_-_44513922 | 0.01 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr12_+_22558928 | 0.00 |
ENSDART00000077548
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr18_-_19016801 | 0.00 |
ENSDART00000129776
|
vwa9
|
von Willebrand factor A domain containing 9 |
| chr3_-_5367142 | 0.00 |
ENSDART00000098927
|
txn2
|
thioredoxin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071887 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) negative regulation of phospholipase activity(GO:0010519) positive regulation of vesicle fusion(GO:0031340) type 2 immune response(GO:0042092) regulation of fatty acid biosynthetic process(GO:0042304) T-helper 2 cell differentiation(GO:0045064) leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.1 | 0.4 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.8 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.1 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.3 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.2 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.1 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.7 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) iridophore differentiation(GO:0050935) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0060333 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) interferon-gamma-mediated signaling pathway(GO:0060333) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0090299 | perichondral ossification(GO:0036073) regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 1.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.2 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |