Project
DANIO-CODE
Navigation
Downloads
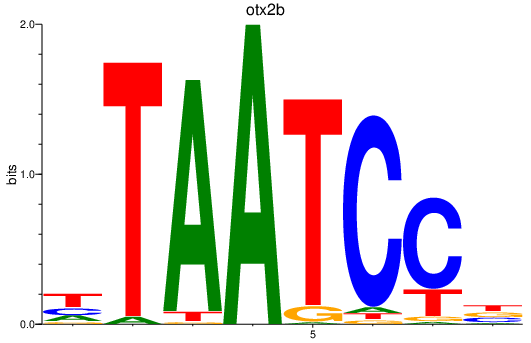
Results for otx2b
Z-value: 1.22
Transcription factors associated with otx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx2b
|
ENSDARG00000011235 | orthodenticle homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx2 | dr10_dc_chr17_-_44133970_44133994 | 0.80 | 2.2e-04 | Click! |
Activity profile of otx2b motif
Sorted Z-values of otx2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of otx2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_26013873 | 9.09 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr19_-_8961733 | 3.45 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr9_-_43178744 | 2.82 |
|
|
|
| chr11_-_29316162 | 2.82 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr1_-_44476084 | 2.66 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr14_-_4166292 | 2.66 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr22_+_30234718 | 2.60 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr5_-_28025315 | 2.57 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr15_-_2689005 | 2.56 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr9_+_26292183 | 2.43 |
|
|
|
| chr7_-_33558939 | 2.43 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr3_+_45375639 | 2.33 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr23_+_35964754 | 2.21 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr15_-_15532980 | 2.20 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_+_67244332 | 2.10 |
ENSDART00000170322
|
rpl13
|
ribosomal protein L13 |
| chr16_+_42925950 | 2.07 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr5_-_28025439 | 2.06 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr23_-_24468191 | 2.04 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr11_-_4216204 | 2.02 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr14_-_36037883 | 1.99 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr8_-_38357549 | 1.98 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr14_-_7774262 | 1.96 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr12_+_6065116 | 1.94 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr5_+_22006860 | 1.92 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr10_+_32160464 | 1.88 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr17_+_16557246 | 1.86 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr10_-_8029671 | 1.85 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr23_+_14058972 | 1.85 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr13_-_39821399 | 1.79 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr15_+_33212930 | 1.78 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr4_-_9172622 | 1.77 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr15_-_18159904 | 1.72 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr14_-_2682064 | 1.72 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_+_24613874 | 1.67 |
ENSDART00000093004
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr15_-_18638643 | 1.58 |
ENSDART00000142010
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr11_-_27455242 | 1.58 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr1_-_10391257 | 1.57 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr10_-_21096635 | 1.55 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr11_-_2133416 | 1.53 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr16_-_41994015 | 1.53 |
|
|
|
| chr25_-_4376129 | 1.52 |
|
|
|
| chr7_+_38123920 | 1.50 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr21_+_21705138 | 1.50 |
ENSDART00000112068
|
defbl1
|
defensin, beta-like 1 |
| chr23_+_2193582 | 1.48 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr19_-_39048324 | 1.48 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr17_+_45931793 | 1.43 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr3_-_32190009 | 1.43 |
|
|
|
| chr7_+_55883471 | 1.42 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr2_-_42011586 | 1.40 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr21_-_20305406 | 1.39 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr20_-_21773202 | 1.38 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr13_-_22901327 | 1.37 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr6_-_13215384 | 1.35 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| chr5_+_58824344 | 1.34 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr11_-_27455348 | 1.33 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr17_-_31147256 | 1.32 |
ENSDART00000055754
|
pkdccb
|
protein kinase domain containing, cytoplasmic b |
| chr10_+_35535209 | 1.32 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr11_-_3841178 | 1.29 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr12_-_22438379 | 1.24 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr7_+_71096788 | 1.23 |
ENSDART00000161871
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr17_+_50622804 | 1.18 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr4_+_5370319 | 1.16 |
ENSDART00000163797
|
ENSDARG00000014395
|
ENSDARG00000014395 |
| chr9_+_7745593 | 1.16 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr14_+_34626233 | 1.15 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr4_-_17226968 | 1.13 |
ENSDART00000177956
ENSDART00000177406 |
BX547928.1
|
ENSDARG00000106175 |
| chr9_-_19154264 | 1.13 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr23_-_28099284 | 1.12 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr7_-_41209891 | 1.12 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr16_+_11260802 | 1.10 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr16_-_33105847 | 1.09 |
|
|
|
| chr23_-_6707246 | 1.09 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr2_+_26273986 | 1.08 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr7_-_13638486 | 1.06 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr17_+_10437343 | 1.06 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr7_-_33559032 | 1.04 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr6_+_36965023 | 1.03 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr8_-_23405325 | 1.02 |
|
|
|
| chr15_-_26703761 | 1.02 |
ENSDART00000087632
|
ENSDARG00000079340
|
ENSDARG00000079340 |
| chr3_+_45375698 | 1.01 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr3_+_23573114 | 1.00 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr7_-_49322226 | 0.98 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr17_-_31041977 | 0.97 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr11_-_16080475 | 0.97 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr15_-_6979997 | 0.95 |
ENSDART00000169944
|
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr23_-_3816492 | 0.95 |
ENSDART00000136394
|
hmga1a
|
high mobility group AT-hook 1a |
| chr1_+_16900546 | 0.95 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr3_+_17947837 | 0.92 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr23_-_35096962 | 0.91 |
ENSDART00000122429
|
klf15
|
Kruppel-like factor 15 |
| chr8_+_14343513 | 0.89 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr17_+_16557298 | 0.88 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr23_-_4175790 | 0.87 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| KN150196v1_+_15525 | 0.86 |
|
|
|
| chr15_+_29183251 | 0.82 |
ENSDART00000153609
|
capns1b
|
calpain, small subunit 1 b |
| chr1_+_50348973 | 0.82 |
|
|
|
| chr7_-_70466713 | 0.81 |
|
|
|
| chr1_+_54403123 | 0.81 |
ENSDART00000152654
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr20_-_46638031 | 0.81 |
ENSDART00000060685
|
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr7_-_28339836 | 0.76 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr11_+_25266623 | 0.76 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr19_-_39048402 | 0.74 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| KN150196v1_+_15459 | 0.74 |
|
|
|
| chr2_-_48317047 | 0.74 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr11_+_30048913 | 0.73 |
|
|
|
| chr5_-_13890202 | 0.72 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr5_+_29327479 | 0.72 |
|
|
|
| chr4_+_15072119 | 0.72 |
|
|
|
| chr15_-_23532475 | 0.70 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr13_-_50299821 | 0.69 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr14_-_9274939 | 0.68 |
|
|
|
| chr6_-_17699493 | 0.68 |
ENSDART00000154180
|
CR388029.1
|
ENSDARG00000097214 |
| chr24_+_37645966 | 0.67 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr5_+_19153421 | 0.66 |
|
|
|
| chr17_+_50622887 | 0.65 |
ENSDART00000134126
|
fermt2
|
fermitin family member 2 |
| chr15_+_46813538 | 0.64 |
|
|
|
| chr21_+_13236509 | 0.64 |
|
|
|
| chr13_-_47510290 | 0.63 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr11_+_2437479 | 0.63 |
ENSDART00000130886
|
NABP2
|
nucleic acid binding protein 2 |
| chr2_-_2456819 | 0.62 |
ENSDART00000013794
ENSDART00000082155 ENSDART00000169871 ENSDART00000167202 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr10_-_18510699 | 0.62 |
|
|
|
| chr13_+_40689329 | 0.61 |
ENSDART00000129532
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr7_+_30698873 | 0.57 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr18_+_10815994 | 0.56 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr17_+_45931756 | 0.56 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr4_-_18447913 | 0.55 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr14_-_29459799 | 0.55 |
ENSDART00000005568
|
pdlim3b
|
PDZ and LIM domain 3b |
| chr2_-_16548451 | 0.54 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr5_+_26217533 | 0.53 |
|
|
|
| chr21_+_5293086 | 0.52 |
|
|
|
| chr7_-_17527827 | 0.50 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr6_+_32428877 | 0.49 |
|
|
|
| chr9_-_28588288 | 0.48 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr1_-_15814788 | 0.48 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr14_+_43599635 | 0.47 |
ENSDART00000155539
|
CR786577.1
|
ENSDARG00000097875 |
| chr18_+_10816190 | 0.46 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr13_+_42276937 | 0.46 |
|
|
|
| chr5_+_42472789 | 0.45 |
ENSDART00000026020
|
wdr54
|
WD repeat domain 54 |
| chr23_-_28099391 | 0.45 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr23_-_28099359 | 0.44 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr7_-_50244215 | 0.42 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr13_-_50299643 | 0.39 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr5_+_69369417 | 0.37 |
|
|
|
| chr10_-_21096686 | 0.35 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr17_-_20959288 | 0.35 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr8_+_27724171 | 0.34 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr3_-_26013660 | 0.34 |
|
|
|
| chr17_+_45932080 | 0.33 |
ENSDART00000157326
|
kif26ab
|
kinesin family member 26Ab |
| chr23_-_28099235 | 0.32 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr7_+_66660781 | 0.32 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr4_-_9172550 | 0.32 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr1_-_37451594 | 0.30 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr20_-_21773327 | 0.29 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_+_25346893 | 0.27 |
|
|
|
| chr11_+_30049412 | 0.24 |
|
|
|
| chr10_+_35610358 | 0.23 |
ENSDART00000077373
|
zdhhc20a
|
zinc finger, DHHC-type containing 20a |
| chr7_-_66641971 | 0.23 |
|
|
|
| chr4_+_11465367 | 0.19 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr13_-_25068717 | 0.19 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr7_-_29871904 | 0.18 |
ENSDART00000173636
|
frmd5
|
FERM domain containing 5 |
| chr2_-_48401640 | 0.16 |
ENSDART00000056277
|
gigfy2
|
GRB10 interacting GYF protein 2 |
| chr11_+_36215760 | 0.16 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr24_-_31257591 | 0.16 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr14_+_14541725 | 0.16 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr18_-_5822255 | 0.15 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr16_+_14820248 | 0.15 |
|
|
|
| chr18_+_10815958 | 0.15 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr20_+_16822321 | 0.14 |
ENSDART00000161226
|
psmc1b
|
proteasome 26S subunit, ATPase 1b |
| chr18_+_7680416 | 0.14 |
ENSDART00000132369
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr14_+_7585570 | 0.12 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr10_-_33212897 | 0.11 |
ENSDART00000081170
|
cux1a
|
cut-like homeobox 1a |
| chr23_-_28368224 | 0.11 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr3_+_24957053 | 0.10 |
ENSDART00000138578
|
st13
|
ST13, Hsp70 interacting protein |
| chr7_+_21365735 | 0.10 |
ENSDART00000173557
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr24_+_3447129 | 0.10 |
ENSDART00000134598
|
wdr37
|
WD repeat domain 37 |
| chr7_-_49322116 | 0.09 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr3_+_21539370 | 0.08 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr17_-_49910160 | 0.07 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr16_-_52090856 | 0.06 |
ENSDART00000155308
|
FO704712.1
|
ENSDARG00000063300 |
| chr11_-_3841149 | 0.05 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr6_-_17699766 | 0.05 |
ENSDART00000170597
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr5_-_27394386 | 0.04 |
ENSDART00000171611
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr7_-_66641207 | 0.03 |
|
|
|
| chr23_+_2193828 | 0.03 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr7_-_50244264 | 0.03 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr1_-_37452112 | 0.02 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr23_-_24468290 | 0.02 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr6_-_13215294 | 0.02 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| KN150196v1_+_15294 | 0.01 |
|
|
|
| chr5_-_68408171 | 0.01 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0045933 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 1.4 | 4.2 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 1.0 | 3.1 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 1.0 | 2.9 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.6 | 1.8 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.6 | 3.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.5 | 2.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 1.3 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.3 | 1.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.3 | 2.8 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 3.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.9 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 1.2 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 1.1 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 0.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 0.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.7 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 2.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 2.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.7 | GO:1902686 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 2.3 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.1 | 1.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 3.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.1 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 3.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0060415 | cardiac muscle tissue morphogenesis(GO:0055008) muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 1.1 | GO:0009416 | response to light stimulus(GO:0009416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.9 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 2.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 1.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 1.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 1.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 4.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |