Project
DANIO-CODE
Navigation
Downloads
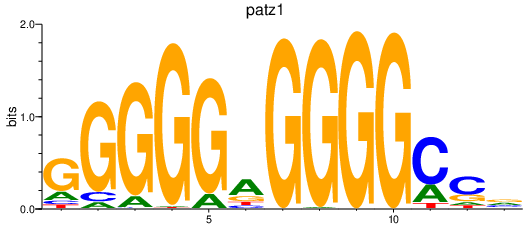
Results for patz1
Z-value: 2.58
Transcription factors associated with patz1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
patz1
|
ENSDARG00000076584 | POZ/BTB and AT hook containing zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| patz1 | dr10_dc_chr6_+_40954138_40954195 | -0.43 | 9.6e-02 | Click! |
Activity profile of patz1 motif
Sorted Z-values of patz1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of patz1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_11913290 | 8.93 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_+_28450576 | 7.43 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr16_-_24220413 | 6.93 |
ENSDART00000103176
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr2_+_11973282 | 6.71 |
ENSDART00000057268
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr8_+_20125687 | 6.62 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr12_-_26760324 | 6.60 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr23_+_10412852 | 6.56 |
ENSDART00000142595
|
krt18
|
keratin 18 |
| chr16_+_17705704 | 6.18 |
|
|
|
| chr7_+_21455138 | 6.18 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr7_+_34417030 | 5.98 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr6_+_47425082 | 5.97 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr15_-_11257445 | 5.82 |
|
|
|
| chr5_+_36487425 | 5.69 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr20_-_25063937 | 5.44 |
ENSDART00000159122
|
epha7
|
eph receptor A7 |
| chr16_-_42486580 | 5.29 |
ENSDART00000148475
|
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr19_+_12066023 | 5.28 |
ENSDART00000130537
|
spag1a
|
sperm associated antigen 1a |
| chr22_+_26543146 | 5.13 |
|
|
|
| chr14_-_943860 | 5.02 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr1_-_674449 | 5.01 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr4_-_25075693 | 4.99 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr6_+_7092350 | 4.99 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr19_-_5338129 | 4.92 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr7_-_23965038 | 4.75 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr5_-_39910235 | 4.72 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr8_+_7301525 | 4.65 |
ENSDART00000092347
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr23_-_2573949 | 4.61 |
ENSDART00000123322
|
snai1b
|
snail family zinc finger 1b |
| chr23_-_24468191 | 4.54 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr12_+_34631232 | 4.53 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr9_+_3458086 | 4.32 |
ENSDART00000160977
ENSDART00000114168 |
CU469503.1
itga6a
|
ENSDARG00000099348 integrin, alpha 6a |
| chr2_+_26523457 | 4.32 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr14_-_46651359 | 4.30 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr14_-_40454194 | 4.28 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_-_21901138 | 4.26 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr6_+_55022668 | 4.25 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr20_-_21773202 | 4.24 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_36647498 | 4.22 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr23_-_29138952 | 4.21 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr16_-_46799406 | 4.18 |
ENSDART00000115244
|
MEX3A
|
mex-3 RNA binding family member A |
| chr5_-_43896815 | 4.09 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr3_-_6078015 | 4.09 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr9_-_48098629 | 4.06 |
|
|
|
| chr8_+_10267246 | 4.06 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr18_-_11216447 | 4.04 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr12_+_34631310 | 4.01 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr15_+_19902697 | 3.96 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr20_-_158899 | 3.91 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr6_+_39373026 | 3.83 |
ENSDART00000157165
|
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr13_-_34667689 | 3.80 |
ENSDART00000025719
|
ism1
|
isthmin 1 |
| chr13_+_11912981 | 3.66 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_-_25693072 | 3.62 |
ENSDART00000122011
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr21_+_5483017 | 3.60 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr7_+_73408688 | 3.58 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr12_+_31501522 | 3.58 |
ENSDART00000144422
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr24_+_5205878 | 3.58 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_+_6054781 | 3.55 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr17_+_8166167 | 3.49 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr3_+_12401750 | 3.48 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr13_+_22173410 | 3.46 |
ENSDART00000173405
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr9_-_44493074 | 3.43 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr24_+_9335428 | 3.41 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr15_-_40328793 | 3.39 |
ENSDART00000155018
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr15_-_4154407 | 3.39 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr7_-_52283383 | 3.39 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr8_+_1122000 | 3.39 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr19_-_24971633 | 3.39 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr22_+_33177432 | 3.38 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr10_+_10830549 | 3.34 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr23_-_7283514 | 3.25 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr5_-_36237656 | 3.24 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr6_+_2849460 | 3.23 |
|
|
|
| chr8_+_25748377 | 3.22 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr10_-_42032702 | 3.19 |
|
|
|
| chr15_+_46114986 | 3.19 |
ENSDART00000156366
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr19_-_39048324 | 3.17 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr16_+_5625301 | 3.15 |
|
|
|
| chr15_-_17074348 | 3.15 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr14_+_33117964 | 3.14 |
ENSDART00000075278
|
atp1b4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr25_-_325847 | 3.14 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr5_+_68410884 | 3.12 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr8_-_14142049 | 3.10 |
ENSDART00000126432
|
rhoaa
|
ras homolog gene family, member Aa |
| chr5_-_25866099 | 3.09 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr13_-_36996246 | 3.07 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr19_+_7234029 | 3.05 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr1_-_50831155 | 3.02 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr8_-_52952226 | 3.02 |
ENSDART00000168185
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr13_+_28601627 | 3.00 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr25_-_340711 | 2.99 |
ENSDART00000178866
|
CU929174.1
|
ENSDARG00000107546 |
| chr11_+_18817797 | 2.94 |
|
|
|
| chr5_+_45822196 | 2.92 |
|
|
|
| chr5_-_43896757 | 2.90 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr8_+_29953399 | 2.89 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr19_-_6384776 | 2.89 |
|
|
|
| chr5_+_66712197 | 2.88 |
ENSDART00000014822
|
ebf2
|
early B-cell factor 2 |
| chr1_-_11591843 | 2.88 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr16_+_29277783 | 2.84 |
ENSDART00000149569
|
CR848047.1
|
ENSDARG00000095906 |
| chr25_+_34270560 | 2.84 |
ENSDART00000146669
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr13_-_34667430 | 2.84 |
ENSDART00000025719
|
ism1
|
isthmin 1 |
| chr7_-_57796486 | 2.81 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr18_+_44656323 | 2.76 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr1_-_51862897 | 2.76 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr9_-_24431684 | 2.73 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr23_+_25782195 | 2.73 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr5_-_31275147 | 2.73 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr19_-_46396376 | 2.69 |
|
|
|
| chr13_+_22350043 | 2.68 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr5_-_31301280 | 2.66 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr15_-_17074107 | 2.66 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr11_+_37345024 | 2.66 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr2_+_3864843 | 2.65 |
ENSDART00000108964
ENSDART00000128514 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr2_-_23348612 | 2.64 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr13_-_14978365 | 2.59 |
|
|
|
| chr3_+_52901602 | 2.59 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr2_+_21342233 | 2.58 |
ENSDART00000062563
|
rreb1b
|
ras responsive element binding protein 1b |
| chr12_-_36138709 | 2.58 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr8_-_53508979 | 2.58 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr14_-_6901415 | 2.57 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr20_-_40422997 | 2.57 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr2_-_44429891 | 2.56 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr19_-_5142310 | 2.56 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr19_-_9603597 | 2.53 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr15_-_33669717 | 2.53 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr4_+_13453879 | 2.50 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr24_-_21114505 | 2.49 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr9_-_22140954 | 2.47 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr22_-_22694468 | 2.46 |
ENSDART00000166794
|
nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr5_+_23585492 | 2.44 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr9_-_33916709 | 2.43 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr1_+_54231447 | 2.39 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr3_-_32190009 | 2.38 |
|
|
|
| chr11_+_23695123 | 2.37 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr11_+_23522743 | 2.36 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr21_+_28408329 | 2.36 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr22_+_24131239 | 2.35 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_-_24602136 | 2.34 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr14_-_9216303 | 2.32 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr8_-_30233470 | 2.31 |
ENSDART00000139864
|
zgc:162939
|
zgc:162939 |
| chr5_-_9497591 | 2.29 |
ENSDART00000113448
|
ENSDARG00000073808
|
ENSDARG00000073808 |
| chr8_+_25235278 | 2.29 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr4_-_16417703 | 2.28 |
ENSDART00000013085
|
dcn
|
decorin |
| chr16_+_17706003 | 2.28 |
|
|
|
| chr11_-_29586549 | 2.27 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr1_-_37990863 | 2.26 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr15_-_1225595 | 2.26 |
|
|
|
| chr24_+_38783264 | 2.26 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| KN149874v1_+_3991 | 2.25 |
|
|
|
| chr23_-_18945009 | 2.25 |
ENSDART00000080064
|
CR847953.1
|
ENSDARG00000057403 |
| chr24_+_35500964 | 2.23 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr16_+_42993841 | 2.21 |
ENSDART00000156767
|
CR854827.4
|
ENSDARG00000097750 |
| chr9_+_56881036 | 2.19 |
|
|
|
| chr7_+_37105282 | 2.18 |
|
|
|
| chr24_+_9335316 | 2.17 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr11_-_40415291 | 2.16 |
|
|
|
| chr17_+_27383737 | 2.13 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr2_+_43619752 | 2.12 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr25_+_4909062 | 2.12 |
ENSDART00000170049
|
parvb
|
parvin, beta |
| chr21_+_13764828 | 2.11 |
ENSDART00000171306
|
stxbp1a
|
syntaxin binding protein 1a |
| chr16_+_5625552 | 2.09 |
|
|
|
| chr10_+_43360137 | 2.07 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr2_-_44330405 | 2.06 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr23_+_6298911 | 2.06 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr16_+_11138924 | 2.06 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr13_-_7897936 | 2.06 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr13_+_4276783 | 2.05 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr18_-_45750 | 2.05 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr21_-_11554253 | 2.05 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr22_-_95158 | 2.01 |
|
|
|
| chr1_-_11591684 | 2.00 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr14_+_1093068 | 1.96 |
ENSDART00000162660
|
si:ch1073-303k11.2
|
si:ch1073-303k11.2 |
| chr16_-_20901879 | 1.95 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr10_+_22759607 | 1.95 |
|
|
|
| chr20_+_545574 | 1.94 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr15_+_42440802 | 1.91 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr18_-_41660821 | 1.90 |
ENSDART00000024087
|
fzd9b
|
frizzled class receptor 9b |
| chr22_-_17652938 | 1.89 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr18_+_14627030 | 1.88 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr16_+_11138879 | 1.87 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr20_+_37940880 | 1.87 |
ENSDART00000165887
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr19_+_4996328 | 1.85 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr21_+_26660833 | 1.84 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr14_+_23986703 | 1.83 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr10_+_31358236 | 1.80 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr19_-_39048402 | 1.80 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr7_-_56493164 | 1.79 |
|
|
|
| chr20_+_20772543 | 1.79 |
ENSDART00000152481
|
rtn1b
|
reticulon 1b |
| chr10_-_21096635 | 1.78 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr5_-_31275259 | 1.77 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr5_+_36014029 | 1.77 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr23_+_22408855 | 1.76 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr16_-_21981065 | 1.75 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr8_+_8763233 | 1.75 |
|
|
|
| chr11_-_80746 | 1.75 |
|
|
|
| chr2_+_47885551 | 1.73 |
|
|
|
| chr9_-_46272322 | 1.72 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| KN149874v1_+_3898 | 1.71 |
|
|
|
| chr15_+_42440952 | 1.70 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr4_-_25075558 | 1.69 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr23_-_624534 | 1.68 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr4_+_149696 | 1.68 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr13_-_17729411 | 1.67 |
ENSDART00000054579
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr5_+_63726719 | 1.67 |
ENSDART00000173886
|
si:ch1073-363c19.3
|
si:ch1073-363c19.3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 1.8 | 5.3 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.6 | 4.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.5 | 4.5 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.3 | 6.7 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 1.2 | 3.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 1.0 | 7.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.9 | 3.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.9 | 3.6 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.8 | 6.7 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.8 | 3.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.7 | 2.0 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.7 | 4.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.7 | 2.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 1.9 | GO:0097037 | heme export(GO:0097037) |
| 0.6 | 3.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.6 | 6.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.6 | 6.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.6 | 2.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 2.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 3.5 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.6 | 4.0 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.5 | 1.5 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.5 | 4.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 3.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.5 | 0.5 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.5 | 14.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.4 | 5.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 3.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.4 | 5.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 1.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.4 | 2.6 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.3 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.3 | 2.4 | GO:0042135 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 1.4 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.3 | 3.4 | GO:0007525 | somatic muscle development(GO:0007525) T-tubule organization(GO:0033292) |
| 0.3 | 5.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.3 | 1.8 | GO:0070293 | renal absorption(GO:0070293) |
| 0.3 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 5.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 3.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 4.0 | GO:0060975 | cardioblast migration(GO:0003260) cardioblast migration to the midline involved in heart field formation(GO:0060975) |
| 0.3 | 3.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.3 | 2.8 | GO:0060021 | palate development(GO:0060021) |
| 0.3 | 4.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.3 | 3.9 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.2 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 2.4 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.2 | 5.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 4.2 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.2 | 4.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 1.4 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 2.0 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.7 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 5.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 3.4 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.2 | 1.3 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 3.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 0.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 2.4 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 2.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 4.1 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 4.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 3.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 5.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 8.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 7.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.2 | 3.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 0.6 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 2.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.1 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 2.0 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 3.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 3.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 3.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 2.3 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 1.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.8 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 1.1 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 4.3 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.1 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.2 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 3.5 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.9 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 3.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 2.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0097178 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.1 | 3.2 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 0.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.2 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 4.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 1.1 | GO:0048883 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 5.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.5 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 1.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.5 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.7 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.7 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 6.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 2.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.7 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 4.2 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 3.4 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 2.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.8 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.4 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.6 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.4 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 1.1 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.2 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.2 | 5.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.5 | 3.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 5.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 4.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 3.6 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.4 | 6.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 1.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 1.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.4 | 5.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 8.0 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.3 | 3.4 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 3.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 2.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 2.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 4.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 8.5 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 6.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 3.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 4.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 11.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 17.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 19.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 2.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 3.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 6.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.4 | GO:0019867 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 5.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 14.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.0 | 7.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.9 | 4.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.8 | 2.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.8 | 3.9 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.8 | 10.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 3.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 1.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.7 | 2.2 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.7 | 2.9 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.7 | 3.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.7 | 2.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.7 | 3.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 6.6 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.5 | 1.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.5 | 5.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.5 | 2.4 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 4.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 9.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 5.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 3.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 3.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 3.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 3.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 2.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 3.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 2.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 2.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 4.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 2.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 4.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 3.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 2.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 5.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.2 | 1.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 10.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 2.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 7.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 2.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.8 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 5.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 4.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 3.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 13.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.6 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 5.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 11.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.2 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.8 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 12.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 6.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.5 | 3.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 6.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 4.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 5.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 1.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 5.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 7.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 2.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 8.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 5.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |