Project
DANIO-CODE
Navigation
Downloads
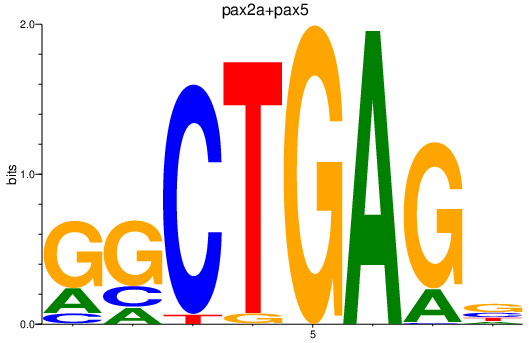
Results for pax2a+pax5
Z-value: 3.36
Transcription factors associated with pax2a+pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2a
|
ENSDARG00000028148 | paired box 2a |
|
pax5
|
ENSDARG00000037383 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax5 | dr10_dc_chr1_+_21033379_21033391 | 0.93 | 1.7e-07 | Click! |
| pax2a | dr10_dc_chr13_+_29640492_29640494 | 0.89 | 3.9e-06 | Click! |
Activity profile of pax2a+pax5 motif
Sorted Z-values of pax2a+pax5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2a+pax5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12033088 | 17.98 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr7_+_29680967 | 14.92 |
ENSDART00000173540
|
tpma
|
alpha-tropomyosin |
| chr3_-_54893038 | 8.32 |
ENSDART00000155871
ENSDART00000109016 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr3_-_48865474 | 7.80 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_-_35928594 | 7.59 |
|
|
|
| chr3_-_34363698 | 7.30 |
|
|
|
| chr2_+_2628865 | 7.28 |
ENSDART00000061955
|
myl13
|
myosin, light chain 13 |
| chr16_+_24063849 | 7.24 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr15_+_32853646 | 7.16 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr13_-_39365252 | 7.02 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr11_+_7518953 | 6.96 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr15_-_23407388 | 6.34 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr23_+_35819625 | 5.89 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr11_-_29316162 | 5.76 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr23_-_18945009 | 5.70 |
ENSDART00000080064
|
CR847953.1
|
ENSDARG00000057403 |
| chr25_+_30827761 | 5.51 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr12_-_17576409 | 5.51 |
ENSDART00000147282
|
pvalb9
|
parvalbumin 9 |
| chr15_-_1858350 | 5.43 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr3_+_32394653 | 5.40 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr16_-_24602919 | 5.37 |
ENSDART00000147478
|
cadm4
|
cell adhesion molecule 4 |
| chr5_+_40722565 | 5.35 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr11_+_30073930 | 5.31 |
ENSDART00000127075
|
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr24_-_40968409 | 5.20 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr7_+_30516734 | 5.19 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr9_-_21420106 | 5.19 |
ENSDART00000102147
|
popdc2
|
popeye domain containing 2 |
| chr8_-_985673 | 5.15 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr21_+_21337628 | 5.15 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr5_+_37053530 | 5.03 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr21_-_23710216 | 5.01 |
ENSDART00000132570
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr20_+_20600211 | 4.99 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr22_+_17091411 | 4.91 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr20_+_582222 | 4.67 |
ENSDART00000152736
ENSDART00000162198 ENSDART00000031759 |
smyd2b
|
SET and MYND domain containing 2b |
| chr23_-_41756325 | 4.67 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr18_-_11216447 | 4.58 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr22_+_18228143 | 4.58 |
ENSDART00000141535
|
BX664610.1
|
ENSDARG00000095557 |
| chr3_-_60973097 | 4.54 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr11_-_80746 | 4.49 |
|
|
|
| chr9_-_34146631 | 4.45 |
ENSDART00000177897
|
AL590134.2
|
ENSDARG00000107869 |
| chr3_+_1413640 | 4.44 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr11_-_22211478 | 4.43 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr24_+_42884 | 4.36 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr13_-_39821399 | 4.34 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr3_+_17210396 | 4.30 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr14_+_37205283 | 4.28 |
ENSDART00000173192
|
pcdh1b
|
protocadherin 1b |
| chr20_-_54192057 | 4.27 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr5_-_36237656 | 4.22 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr16_+_54000890 | 4.21 |
ENSDART00000014098
ENSDART00000150190 |
ggctb
|
gamma-glutamylcyclotransferase b |
| chr24_-_41448993 | 4.21 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr10_-_25695574 | 4.20 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr5_-_67233396 | 4.13 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr5_+_26925392 | 4.10 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_32605735 | 4.08 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr22_-_18087170 | 4.07 |
|
|
|
| chr5_+_26925238 | 4.06 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr19_-_3299610 | 4.06 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr5_+_26832823 | 4.05 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr8_-_19364689 | 4.04 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr19_-_29205158 | 4.03 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr5_-_25866099 | 4.01 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr14_-_36037883 | 3.99 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr6_-_32718634 | 3.98 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr3_-_55956030 | 3.97 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr15_+_548292 | 3.94 |
ENSDART00000178563
|
ftr86
|
finTRIM family, member 86 |
| chr8_+_53118919 | 3.93 |
|
|
|
| chr20_+_25441689 | 3.92 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr25_-_31218193 | 3.91 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr23_-_27226280 | 3.90 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr8_-_40231417 | 3.89 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr7_-_71958596 | 3.83 |
|
|
|
| chr19_+_38834972 | 3.82 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr20_+_15119754 | 3.82 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr7_+_31608828 | 3.80 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_+_30681170 | 3.80 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr6_+_49096966 | 3.79 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr3_+_39425125 | 3.77 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr5_-_55843553 | 3.76 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr9_-_12063411 | 3.75 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr5_-_34016383 | 3.72 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr9_-_7560915 | 3.72 |
ENSDART00000081553
|
desma
|
desmin a |
| chr18_-_45750 | 3.72 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_+_24046789 | 3.63 |
|
|
|
| chr6_+_52791515 | 3.62 |
ENSDART00000065682
|
matn4
|
matrilin 4 |
| chr6_-_23455346 | 3.58 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr24_-_34449257 | 3.57 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr18_+_9213713 | 3.56 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr7_+_58718614 | 3.55 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr7_+_11148688 | 3.55 |
ENSDART00000114383
|
mesdc1
|
mesoderm development candidate 1 |
| chr24_-_37680649 | 3.53 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr11_+_3186396 | 3.52 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr6_-_8126955 | 3.52 |
ENSDART00000132412
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr11_+_43083105 | 3.49 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr2_-_15894180 | 3.47 |
ENSDART00000172588
|
col11a1b
|
collagen, type XI, alpha 1b |
| KN149874v1_+_3991 | 3.46 |
|
|
|
| chr16_+_4288700 | 3.46 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr22_+_223797 | 3.43 |
|
|
|
| chr15_+_7068253 | 3.42 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr23_-_21520413 | 3.39 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr25_-_7346766 | 3.36 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr13_+_24531753 | 3.36 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr14_+_23980348 | 3.34 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr8_+_26273354 | 3.34 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr2_-_57309939 | 3.34 |
|
|
|
| chr13_+_22545318 | 3.32 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr7_+_19765237 | 3.31 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr16_+_29109050 | 3.30 |
ENSDART00000122681
|
CR848841.1
|
Nestin |
| chr13_-_49826330 | 3.30 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr1_+_53259374 | 3.28 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr19_+_38834765 | 3.28 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr1_+_53353690 | 3.28 |
ENSDART00000126339
|
dla
|
deltaA |
| chr9_-_2588401 | 3.27 |
ENSDART00000161018
|
sp9
|
sp9 transcription factor |
| chr1_-_28241500 | 3.26 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr2_+_22324846 | 3.25 |
ENSDART00000044371
|
tox
|
thymocyte selection-associated high mobility group box |
| chr1_-_51862897 | 3.23 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_-_37587227 | 3.22 |
ENSDART00000173552
|
adcy7
|
adenylate cyclase 7 |
| chr6_+_2849460 | 3.18 |
|
|
|
| chr1_-_5048203 | 3.18 |
ENSDART00000150863
ENSDART00000163417 |
nrp2a
|
neuropilin 2a |
| chr2_+_27344969 | 3.17 |
ENSDART00000140059
|
BX511023.1
|
ENSDARG00000092372 |
| chr24_+_4341242 | 3.17 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr13_+_17541579 | 3.16 |
ENSDART00000137776
|
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr5_-_28025315 | 3.16 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr13_-_42630425 | 3.16 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr2_+_30932612 | 3.16 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr1_-_7456961 | 3.15 |
ENSDART00000152295
|
FAM83G
|
family with sequence similarity 83 member G |
| KN149874v1_+_3898 | 3.10 |
|
|
|
| chr10_-_24422257 | 3.07 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr16_+_1227270 | 3.06 |
|
|
|
| chr2_+_27344633 | 3.06 |
ENSDART00000178275
|
cdh7
|
cadherin 7, type 2 |
| chr9_+_308260 | 3.06 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr23_-_9924987 | 3.05 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr10_+_26020615 | 3.05 |
ENSDART00000128292
ENSDART00000170275 ENSDART00000166164 ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr16_-_54634679 | 3.03 |
ENSDART00000075275
ENSDART00000115024 |
pklr
|
pyruvate kinase, liver and RBC |
| chr13_+_7575753 | 3.03 |
|
|
|
| chr2_-_59293269 | 3.02 |
ENSDART00000148286
|
ftr39p
|
finTRIM family, member 39, pseudogene |
| chr16_+_28661916 | 3.01 |
ENSDART00000046209
|
acbd7
|
acyl-CoA binding domain containing 7 |
| chr25_+_32707410 | 2.99 |
ENSDART00000125892
ENSDART00000121680 |
ENSDARG00000087402
|
ENSDARG00000087402 |
| chr2_+_49358871 | 2.96 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr8_+_47815343 | 2.96 |
ENSDART00000053285
|
ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 |
| chr6_+_39362786 | 2.95 |
ENSDART00000151322
|
ENSDARG00000028618
|
ENSDARG00000028618 |
| chr20_+_10502210 | 2.94 |
ENSDART00000155888
|
AL845550.1
|
ENSDARG00000097342 |
| chr10_-_36462538 | 2.94 |
ENSDART00000161823
|
ubl3a
|
ubiquitin-like 3a |
| chr23_+_23730717 | 2.94 |
|
|
|
| chr18_-_6494014 | 2.93 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr17_+_32550616 | 2.91 |
|
|
|
| chr4_+_21319791 | 2.90 |
|
|
|
| chr15_+_7068009 | 2.90 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr16_+_5466342 | 2.87 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr19_-_35642440 | 2.86 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr4_-_12862869 | 2.86 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr7_+_34394526 | 2.86 |
|
|
|
| chr16_+_55167489 | 2.85 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr9_-_34129128 | 2.85 |
|
|
|
| chr17_-_32913432 | 2.83 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr6_-_24003717 | 2.83 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr3_-_58590651 | 2.83 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr21_-_3301751 | 2.82 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr5_-_62747812 | 2.80 |
|
|
|
| chr11_-_35501227 | 2.73 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr15_-_39756101 | 2.72 |
|
|
|
| chr20_+_26408687 | 2.70 |
|
|
|
| chr24_+_40625215 | 2.70 |
|
|
|
| chr9_+_25964943 | 2.70 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr23_+_37754409 | 2.69 |
|
|
|
| chr11_+_25022419 | 2.68 |
|
|
|
| chr7_+_20046425 | 2.68 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr23_-_27226387 | 2.67 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr3_-_2686533 | 2.67 |
|
|
|
| chr11_+_37639045 | 2.67 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr22_-_37417903 | 2.67 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr21_+_20512024 | 2.67 |
ENSDART00000126005
|
efna5a
|
ephrin-A5a |
| chr3_-_60929921 | 2.65 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr2_-_40170303 | 2.65 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr7_+_31608878 | 2.65 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_+_29640492 | 2.65 |
ENSDART00000160944
|
pax2a
|
paired box 2a |
| chr3_+_24327586 | 2.64 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr14_+_28136958 | 2.63 |
|
|
|
| chr11_-_18542803 | 2.63 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr4_+_5767840 | 2.63 |
ENSDART00000076494
|
ngs
|
notochord granular surface |
| chr23_+_36832325 | 2.62 |
|
|
|
| chr23_+_27749014 | 2.62 |
ENSDART00000128833
|
rps26
|
ribosomal protein S26 |
| chr5_-_68145952 | 2.61 |
ENSDART00000141917
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr14_-_46648563 | 2.61 |
|
|
|
| chr5_-_28025439 | 2.58 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr7_-_31170266 | 2.58 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr11_+_36933891 | 2.56 |
ENSDART00000170542
|
ENSDARG00000100406
|
ENSDARG00000100406 |
| chr14_-_32838287 | 2.55 |
|
|
|
| chr9_+_24089828 | 2.54 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr6_-_39053400 | 2.54 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr4_-_12862708 | 2.53 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr11_-_23226859 | 2.53 |
ENSDART00000159221
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr21_-_5852663 | 2.53 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr13_-_31516399 | 2.52 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr19_+_42183946 | 2.52 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr7_-_18894639 | 2.51 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
| chr17_+_53224434 | 2.51 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr23_+_36032206 | 2.49 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr3_-_54884533 | 2.49 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr10_+_16111842 | 2.49 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr21_-_28883441 | 2.49 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr17_+_25503946 | 2.49 |
|
|
|
| chr5_-_7134405 | 2.49 |
ENSDART00000177935
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.8 | 7.2 | GO:0055016 | hypochord development(GO:0055016) |
| 1.8 | 5.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 1.6 | 6.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.6 | 6.3 | GO:0061626 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 1.5 | 6.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.4 | 4.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 1.3 | 5.4 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 1.3 | 4.0 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 1.3 | 3.8 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 1.2 | 6.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.2 | 5.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 1.2 | 3.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 1.2 | 3.5 | GO:0097435 | fibril organization(GO:0097435) |
| 1.1 | 3.3 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 1.1 | 3.3 | GO:0051580 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 1.0 | 29.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 1.0 | 10.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.9 | 2.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.9 | 5.4 | GO:0072505 | divalent inorganic anion homeostasis(GO:0072505) |
| 0.9 | 3.5 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.8 | 2.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.8 | 2.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.8 | 1.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.8 | 2.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.8 | 3.1 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.8 | 4.7 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.8 | 2.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.8 | 2.3 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.7 | 8.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.7 | 4.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.7 | 2.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) cellular response to vitamin D(GO:0071305) |
| 0.7 | 8.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.7 | 2.0 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.7 | 3.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.6 | 10.9 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.6 | 1.9 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.6 | 1.9 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.6 | 4.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.6 | 8.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 1.6 | GO:0097186 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.5 | 3.3 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.5 | 1.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.5 | 2.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.5 | 2.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.5 | 4.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.5 | 2.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.5 | 2.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.5 | 4.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.5 | 2.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.5 | 0.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.5 | 5.4 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.5 | 2.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.5 | 4.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.5 | 3.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.4 | 1.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 5.2 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.4 | 3.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 5.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 1.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 7.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 12.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 1.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 6.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.4 | 24.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.4 | 1.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.3 | 2.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 1.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 2.3 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.3 | 1.9 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 2.3 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.9 | GO:0070293 | renal absorption(GO:0070293) |
| 0.3 | 1.9 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 2.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 1.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 0.9 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.3 | 0.6 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.3 | 1.8 | GO:0032371 | membrane raft organization(GO:0031579) regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.3 | 4.4 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.3 | 5.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 0.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.3 | 5.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 1.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 2.6 | GO:0060035 | notochord cell development(GO:0060035) |
| 0.3 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 2.0 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.3 | 3.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 0.8 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.3 | 0.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 5.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.5 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.2 | 4.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 3.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.2 | 1.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 0.9 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.2 | 5.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 4.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.6 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 | 0.7 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 3.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 1.6 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.2 | 2.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.9 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 7.0 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.2 | 1.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 0.6 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.2 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 2.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.8 | GO:0061032 | pericardium development(GO:0060039) visceral serous pericardium development(GO:0061032) |
| 0.2 | 1.6 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.2 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 0.8 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.2 | 2.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.4 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 2.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 8.9 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.2 | 4.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.2 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 7.6 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.2 | 2.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 3.9 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.2 | 0.5 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 4.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.2 | 3.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 0.7 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 2.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.2 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 9.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 3.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.4 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 2.0 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 2.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 2.8 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 1.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 4.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 3.4 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 5.7 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.1 | 12.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 2.3 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.1 | 0.2 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.7 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 2.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.9 | GO:0022408 | negative regulation of cell-cell adhesion(GO:0022408) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 20.2 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 4.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 2.3 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 5.5 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 3.0 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.1 | 0.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 2.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 2.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 3.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 4.4 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.4 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 1.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 2.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 2.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 1.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.4 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.1 | 5.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 0.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.8 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.2 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.9 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 2.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.4 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 0.0 | 4.2 | GO:0008015 | blood circulation(GO:0008015) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.4 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.3 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 3.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.7 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 1.4 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 4.0 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.0 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 1.5 | 10.8 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 1.3 | 28.2 | GO:0005861 | troponin complex(GO:0005861) |
| 1.2 | 3.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.0 | 5.0 | GO:0008091 | spectrin(GO:0008091) |
| 1.0 | 4.0 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.9 | 10.1 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 2.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.7 | 5.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 3.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.6 | 2.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.6 | 6.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.5 | 2.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.5 | 1.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.4 | 2.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 3.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 2.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.4 | 2.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 1.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 8.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 2.3 | GO:0005903 | brush border(GO:0005903) |
| 0.3 | 2.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 1.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 1.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 2.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 10.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 9.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 11.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 2.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 16.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 2.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 12.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 5.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 6.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 7.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 6.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.7 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 3.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 13.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 3.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0030427 | growth cone(GO:0030426) site of polarized growth(GO:0030427) |
| 0.1 | 1.3 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) cell cortex region(GO:0099738) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 3.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 40.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.2 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 2.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 38.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 6.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 1.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 3.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 4.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 10.7 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 4.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.9 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) |
| 0.0 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 5.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 25.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.5 | 10.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.2 | 3.6 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 1.2 | 4.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.1 | 3.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.0 | 3.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.0 | 4.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 1.0 | 7.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 4.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 2.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.8 | 2.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.8 | 3.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 7.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 6.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.7 | 2.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.6 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.5 | 2.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 4.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.5 | 4.9 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.5 | 2.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.5 | 5.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 2.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 4.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 3.5 | GO:0008199 | acid phosphatase activity(GO:0003993) ferric iron binding(GO:0008199) |
| 0.4 | 4.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 3.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.4 | 2.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.4 | 3.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 2.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 5.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 6.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.5 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.4 | 3.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 1.4 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.4 | 3.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 4.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 2.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.4 | 1.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 2.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 4.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 5.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 1.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 1.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 6.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.3 | 1.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 4.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.3 | 13.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 18.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 4.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 2.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 1.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 7.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 5.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 0.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.3 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 4.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 8.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 1.7 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.2 | 2.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 3.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 3.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.2 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 1.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 16.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.2 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 20.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.2 | 2.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 3.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 4.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 8.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.2 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.1 | 13.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 0.4 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 3.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 49.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.7 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 6.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.1 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 9.4 | GO:0019900 | kinase binding(GO:0019900) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 5.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 55.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 4.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 1.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 10.7 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 5.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 3.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 5.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 7.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 5.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 2.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 8.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 2.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 2.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 2.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 5.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 6.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 3.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 5.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 3.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.6 | 5.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 7.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 4.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 7.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 2.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 3.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 3.9 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 1.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.7 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 2.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 12.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 7.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 0.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |