Project
DANIO-CODE
Navigation
Downloads
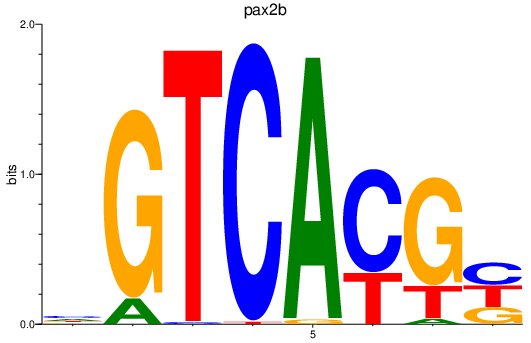
Results for pax2b
Z-value: 1.85
Transcription factors associated with pax2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2b
|
ENSDARG00000032578 | paired box 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2b | dr10_dc_chr12_-_45714172_45714180 | -0.75 | 8.2e-04 | Click! |
Activity profile of pax2b motif
Sorted Z-values of pax2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_23704663 | 4.67 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr8_+_52456064 | 4.21 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr19_+_15536640 | 4.06 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_25281122 | 3.55 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr8_-_2557556 | 3.45 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr10_+_16543480 | 3.24 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr24_-_24999240 | 3.23 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr23_-_22596648 | 3.10 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr7_-_55346967 | 2.74 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr6_+_135002 | 2.64 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr11_+_12983670 | 2.60 |
ENSDART00000166126
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr15_+_29092022 | 2.60 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr16_+_9823111 | 2.58 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr23_-_35384257 | 2.57 |
ENSDART00000113643
|
fbxo25
|
F-box protein 25 |
| chr11_-_44539778 | 2.49 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr10_-_2944190 | 2.47 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr3_-_20912934 | 2.45 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr4_+_16020464 | 2.44 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr19_+_31997954 | 2.43 |
ENSDART00000164108
|
gmnn
|
geminin, DNA replication inhibitor |
| chr3_-_32231069 | 2.43 |
ENSDART00000035545
|
prmt1
|
protein arginine methyltransferase 1 |
| chr17_+_52526741 | 2.37 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr13_+_35799681 | 2.36 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_+_35799602 | 2.33 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr3_-_15325357 | 2.25 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr21_-_3548863 | 2.24 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr16_-_55211053 | 2.21 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr1_-_55072271 | 2.19 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr2_-_57339717 | 2.14 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr2_+_10858480 | 2.13 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr14_+_24543399 | 2.12 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr5_-_31257158 | 2.11 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr22_+_8856370 | 2.05 |
ENSDART00000178799
|
CABZ01046433.1
|
ENSDARG00000109049 |
| chr16_+_9822930 | 2.05 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr10_+_36718872 | 2.02 |
ENSDART00000063359
|
ucp3
|
uncoupling protein 3 |
| chr6_-_53145464 | 1.96 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr14_+_19859164 | 1.96 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr23_+_33792145 | 1.96 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr8_+_28715451 | 1.95 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr22_+_37688839 | 1.93 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr10_-_2944295 | 1.93 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr17_+_25168837 | 1.92 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr1_+_45015350 | 1.91 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr11_+_13118866 | 1.90 |
ENSDART00000123257
|
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr10_-_45182378 | 1.87 |
ENSDART00000161815
|
polm
|
polymerase (DNA directed), mu |
| chr1_+_5437327 | 1.86 |
ENSDART00000092324
|
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr24_-_24999067 | 1.83 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_19837266 | 1.83 |
ENSDART00000015780
|
ercc4
|
excision repair cross-complementation group 4 |
| chr5_-_47609402 | 1.83 |
|
|
|
| chr8_+_40236694 | 1.80 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr5_-_28931727 | 1.79 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr4_-_951763 | 1.78 |
ENSDART00000093289
|
xrcc6bp1
|
XRCC6 binding protein 1 |
| chr22_+_22413803 | 1.78 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr15_+_29091983 | 1.76 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr12_-_926596 | 1.76 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr3_+_42217236 | 1.75 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr17_+_25163592 | 1.74 |
ENSDART00000100277
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr2_+_15379961 | 1.73 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr5_-_9036049 | 1.73 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr5_-_47609309 | 1.73 |
|
|
|
| chr10_-_24796421 | 1.72 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr6_-_8468819 | 1.71 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr22_-_52486 | 1.70 |
|
|
|
| chr19_+_2942485 | 1.69 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr18_+_44775449 | 1.68 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr5_-_14000166 | 1.67 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr2_+_15379717 | 1.67 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr3_+_32700459 | 1.66 |
ENSDART00000132679
ENSDART00000035759 |
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr22_-_25592743 | 1.65 |
ENSDART00000110638
ENSDART00000172092 |
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr3_+_16572387 | 1.64 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr23_-_24416864 | 1.64 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr16_+_53632289 | 1.63 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr6_-_10676857 | 1.62 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr13_-_7972442 | 1.62 |
ENSDART00000128005
|
atl2
|
atlastin GTPase 2 |
| chr10_+_16543577 | 1.62 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr8_+_40236654 | 1.61 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr10_+_18953548 | 1.61 |
ENSDART00000030205
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr23_-_24416911 | 1.59 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr10_-_44441197 | 1.59 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr15_+_29092224 | 1.57 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr16_-_13723352 | 1.57 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr13_-_7972530 | 1.56 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr14_+_21373857 | 1.55 |
ENSDART00000079649
ENSDART00000165795 |
ndufs8b
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8b |
| chr14_+_29932533 | 1.54 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr8_-_2557506 | 1.53 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr21_+_7737161 | 1.53 |
|
|
|
| chr3_+_56882443 | 1.51 |
|
|
|
| chr5_-_9035789 | 1.50 |
ENSDART00000124384
|
gak
|
cyclin G associated kinase |
| chr19_+_791656 | 1.49 |
ENSDART00000138406
|
tmem79a
|
transmembrane protein 79a |
| chr5_-_40503137 | 1.48 |
ENSDART00000051070
|
golph3
|
golgi phosphoprotein 3 |
| chr11_-_14044356 | 1.48 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr18_+_6339577 | 1.48 |
ENSDART00000136333
|
wash1
|
WAS protein family homolog 1 |
| chr20_+_30476065 | 1.47 |
ENSDART00000062441
|
rnaseh1
|
ribonuclease H1 |
| chr12_-_30244575 | 1.46 |
ENSDART00000152981
|
tdrd1
|
tudor domain containing 1 |
| chr8_-_40236572 | 1.46 |
ENSDART00000053158
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr3_+_16772351 | 1.46 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr3_-_26060787 | 1.46 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr6_+_18894174 | 1.46 |
ENSDART00000165806
|
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr9_-_11616001 | 1.44 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr7_+_28905795 | 1.43 |
ENSDART00000146655
|
aph1b
|
APH1B gamma secretase subunit |
| chr20_-_53835111 | 1.40 |
ENSDART00000177147
|
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr21_+_22364359 | 1.40 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr5_-_18502344 | 1.40 |
ENSDART00000137022
ENSDART00000090494 ENSDART00000165701 |
golga3
|
golgin A3 |
| chr22_-_52569 | 1.39 |
|
|
|
| chr21_-_19883182 | 1.39 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr13_+_45387478 | 1.38 |
ENSDART00000019113
|
tmem57b
|
transmembrane protein 57b |
| chr12_+_19837298 | 1.37 |
ENSDART00000015780
|
ercc4
|
excision repair cross-complementation group 4 |
| chr2_+_15380054 | 1.37 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr10_+_42085689 | 1.36 |
ENSDART00000009227
|
orai1b
|
ORAI calcium release-activated calcium modulator 1b |
| chr8_+_50202152 | 1.34 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr7_-_9628384 | 1.33 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr5_-_11530803 | 1.33 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr10_-_44441390 | 1.33 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr7_+_66410533 | 1.32 |
ENSDART00000027616
ENSDART00000162763 |
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr7_+_27184121 | 1.32 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr8_-_22521204 | 1.32 |
ENSDART00000134542
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr18_-_3455211 | 1.31 |
ENSDART00000163762
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr2_+_10858655 | 1.31 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr6_-_51811024 | 1.31 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr23_+_17583666 | 1.30 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr10_+_16544000 | 1.30 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr25_-_13394261 | 1.30 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr7_+_22027528 | 1.30 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr3_+_33608070 | 1.29 |
|
|
|
| chr6_-_8225364 | 1.29 |
|
|
|
| chr24_+_22614373 | 1.29 |
ENSDART00000135392
|
si:dkey-7n6.2
|
si:dkey-7n6.2 |
| chr16_+_9822879 | 1.29 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr16_-_13723295 | 1.28 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr23_+_17583383 | 1.28 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr19_-_27239547 | 1.28 |
ENSDART00000149132
|
neu1
|
neuraminidase 1 |
| chr11_+_44738699 | 1.28 |
ENSDART00000172301
ENSDART00000170118 |
tk1
|
thymidine kinase 1, soluble |
| chr13_-_50149031 | 1.27 |
|
|
|
| chr3_+_9504996 | 1.27 |
ENSDART00000171467
|
si:dkey-29p9.3
|
si:dkey-29p9.3 |
| chr15_+_45576378 | 1.26 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr15_-_23786063 | 1.26 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr14_+_29601046 | 1.25 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| KN149698v1_-_88818 | 1.25 |
|
|
|
| chr22_+_25059272 | 1.24 |
ENSDART00000177284
|
rrbp1b
|
ribosome binding protein 1b |
| chr25_+_30627892 | 1.24 |
|
|
|
| chr6_-_8468961 | 1.24 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr4_+_25923205 | 1.24 |
ENSDART00000136927
|
vetz
|
vezatin, adherens junctions transmembrane protein |
| chr22_-_26537845 | 1.23 |
|
|
|
| chr9_-_9864149 | 1.23 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr11_-_44738541 | 1.23 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr13_-_31267133 | 1.23 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr13_-_25620394 | 1.23 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr2_+_8314513 | 1.23 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr6_+_135255 | 1.23 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr20_+_54544851 | 1.23 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr22_+_37689055 | 1.21 |
|
|
|
| chr13_-_35766296 | 1.21 |
ENSDART00000162399
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr20_-_26161022 | 1.21 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr19_+_14490203 | 1.21 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr17_+_44327269 | 1.21 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr16_+_26858575 | 1.21 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr6_+_49724436 | 1.20 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr7_+_71262845 | 1.20 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr5_-_69171508 | 1.19 |
ENSDART00000127782
|
ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr20_-_40470331 | 1.18 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr10_-_24754849 | 1.17 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr3_-_58400345 | 1.17 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr25_-_13774670 | 1.16 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr11_-_30971444 | 1.16 |
ENSDART00000170700
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr7_+_24119940 | 1.16 |
ENSDART00000108753
|
poln
|
polymerase (DNA directed) nu |
| chr7_+_46970462 | 1.16 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr15_-_14102102 | 1.15 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr1_-_54488824 | 1.15 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr16_+_53632152 | 1.15 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr19_+_34263554 | 1.14 |
ENSDART00000161290
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr4_-_16846903 | 1.14 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr5_-_14000289 | 1.13 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr4_+_9357791 | 1.13 |
|
|
|
| chr21_+_20350152 | 1.12 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr8_-_31706973 | 1.12 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr8_+_50201986 | 1.11 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr21_+_34053739 | 1.11 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr7_-_41532268 | 1.11 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr17_+_49995001 | 1.10 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr25_-_17282381 | 1.10 |
ENSDART00000178173
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr11_-_31012985 | 1.09 |
ENSDART00000162605
|
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr19_+_14295978 | 1.09 |
ENSDART00000168260
|
nudc
|
nudC nuclear distribution protein |
| chr8_+_40236817 | 1.09 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr6_+_38882608 | 1.08 |
ENSDART00000019939
|
bin2b
|
bridging integrator 2b |
| chr3_-_29837017 | 1.08 |
ENSDART00000139310
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr24_-_21758198 | 1.08 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr4_-_7867294 | 1.08 |
ENSDART00000167943
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr14_+_29601073 | 1.07 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr3_-_54245542 | 1.07 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr5_+_56896961 | 1.07 |
|
|
|
| chr16_+_17762256 | 1.06 |
ENSDART00000128672
|
tmem238b
|
transmembrane protein 238b |
| chr6_-_41141242 | 1.06 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr9_-_21729953 | 1.06 |
|
|
|
| chr20_-_223222 | 1.06 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr19_-_35400339 | 1.05 |
|
|
|
| chr23_-_31986679 | 1.05 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_15891060 | 1.05 |
ENSDART00000154181
|
syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr13_-_23825787 | 1.05 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr13_+_40566900 | 1.04 |
ENSDART00000057047
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr5_-_66687335 | 1.04 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr18_+_6417959 | 1.04 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr9_+_34571082 | 1.04 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:1990379 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.9 | 5.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.8 | 2.5 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.7 | 2.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.7 | 2.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.7 | 3.6 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.7 | 2.1 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.7 | 2.8 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.7 | 2.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.6 | 3.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.6 | 2.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 2.8 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) valine biosynthetic process(GO:0009099) |
| 0.5 | 1.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.5 | 1.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.5 | 5.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 2.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.4 | 1.3 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.4 | 0.4 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.4 | 2.0 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.4 | 3.6 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.4 | 2.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.4 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 9.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 1.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 5.9 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.3 | 1.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.3 | 0.9 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.2 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.8 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 0.9 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 1.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.3 | 0.8 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.3 | 7.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 2.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.7 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 6.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.2 | 3.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 0.9 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 1.5 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 0.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 0.8 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 2.0 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 2.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 2.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 2.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.2 | 0.6 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 0.6 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 1.3 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 1.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 1.0 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 2.0 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.2 | 1.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.5 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 3.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 2.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.2 | 2.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 0.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 2.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 4.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.8 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 5.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.4 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.1 | 0.5 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 1.0 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 4.9 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 1.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.4 | GO:1903335 | regulation of vacuolar transport(GO:1903335) |
| 0.1 | 0.3 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.8 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 | 0.7 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 2.6 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.3 | GO:0001715 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.2 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.6 | GO:1901314 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.1 | 3.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.2 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 | 2.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.0 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 1.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 1.3 | GO:0006213 | pyrimidine nucleoside metabolic process(GO:0006213) |
| 0.1 | 1.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) RNA 5'-end processing(GO:0000966) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.8 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 3.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 0.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.5 | GO:0072665 | protein localization to vacuole(GO:0072665) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle elongation(GO:0000022) positive regulation of cytokinesis(GO:0032467) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 6.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.7 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.0 | 1.4 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.8 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.9 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.2 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 1.3 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.8 | GO:0007281 | germ cell development(GO:0007281) |
| 0.0 | 1.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.4 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 1.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.6 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 1.1 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.0 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 2.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.7 | 2.0 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.6 | 7.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 3.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 3.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 2.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 7.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 1.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 1.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.3 | 1.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 3.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.6 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 3.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0045178 | basal plasma membrane(GO:0009925) basal part of cell(GO:0045178) |
| 0.1 | 4.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 10.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 1.0 | 5.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.9 | 6.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 2.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.7 | 3.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.6 | 1.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.6 | 2.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.5 | 2.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 5.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 5.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 2.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 1.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 3.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 1.5 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.4 | 1.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 1.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 2.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.4 | 1.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 2.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.3 | 2.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 0.9 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 3.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.3 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.0 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 3.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.7 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 0.7 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.2 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 1.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.0 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.2 | 0.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 5.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 3.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.7 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 1.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 2.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 0.5 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 0.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 5.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.4 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 1.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.8 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.5 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.1 | 2.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.6 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 5.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0019869 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 5.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 2.4 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.1 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 2.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 2.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 1.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 5.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 0.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 1.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.7 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.1 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |