Project
DANIO-CODE
Navigation
Downloads
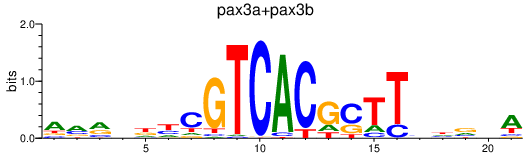
Results for pax3a+pax3b
Z-value: 1.45
Transcription factors associated with pax3a+pax3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax3a
|
ENSDARG00000010192 | paired box 3a |
|
pax3b
|
ENSDARG00000028348 | paired box 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax3b | dr10_dc_chr15_-_42250788_42250820 | -0.94 | 3.8e-08 | Click! |
| pax3a | dr10_dc_chr2_-_47578695_47578757 | -0.93 | 1.7e-07 | Click! |
Activity profile of pax3a+pax3b motif
Sorted Z-values of pax3a+pax3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax3a+pax3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_24249672 | 7.16 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr12_+_22551105 | 5.79 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr10_-_34971985 | 5.53 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr8_-_410190 | 4.38 |
ENSDART00000151155
|
trim36
|
tripartite motif containing 36 |
| chr6_-_53145464 | 4.23 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr7_-_24249630 | 3.71 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr12_-_14104939 | 3.53 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr1_+_11508857 | 3.45 |
ENSDART00000172334
ENSDART00000112171 |
tdrd7a
|
tudor domain containing 7 a |
| chr14_-_21320696 | 3.43 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr7_-_24724911 | 3.21 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr22_+_8856370 | 3.17 |
ENSDART00000178799
|
CABZ01046433.1
|
ENSDARG00000109049 |
| chr16_+_9822930 | 3.15 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr5_+_21902777 | 3.10 |
ENSDART00000141385
|
si:dkey-27p18.3
|
si:dkey-27p18.3 |
| chr16_+_9823111 | 2.77 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr8_-_410293 | 2.55 |
ENSDART00000164886
|
trim36
|
tripartite motif containing 36 |
| chr7_+_27184121 | 2.51 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr10_-_34971926 | 2.51 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr20_-_36510718 | 2.40 |
|
|
|
| chr25_-_13394261 | 2.29 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr22_+_62610 | 2.22 |
ENSDART00000062264
|
sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_2615745 | 2.21 |
ENSDART00000160777
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr10_+_16543480 | 2.20 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr6_-_53145335 | 2.15 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr6_-_53145582 | 2.03 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr1_+_50893368 | 2.01 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr4_+_4825461 | 1.98 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr22_-_760140 | 1.96 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr20_-_18482817 | 1.86 |
ENSDART00000159636
|
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr2_-_4191540 | 1.80 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr1_+_25911314 | 1.77 |
ENSDART00000011645
|
coro2a
|
coronin, actin binding protein, 2A |
| chr1_+_57878303 | 1.77 |
ENSDART00000162460
|
si:ch73-221f6.3
|
si:ch73-221f6.3 |
| chr16_+_9822879 | 1.77 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr21_-_19883182 | 1.74 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_-_20940726 | 1.72 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr24_-_20063951 | 1.68 |
ENSDART00000139329
|
cry-dash
|
cryptochrome DASH |
| chr10_+_38720500 | 1.67 |
ENSDART00000067448
|
acat1
|
acetyl-CoA acetyltransferase 1 |
| chr12_+_18849010 | 1.65 |
ENSDART00000078494
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr8_-_47215582 | 1.65 |
ENSDART00000147606
|
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr1_+_45907424 | 1.65 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr21_-_2261849 | 1.62 |
ENSDART00000172041
ENSDART00000161554 |
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr2_-_47766563 | 1.59 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr12_+_18848952 | 1.59 |
ENSDART00000153235
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr4_-_7861030 | 1.56 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr21_-_19882766 | 1.51 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr7_+_20091996 | 1.49 |
ENSDART00000138786
|
ponzr1
|
plac8 onzin related protein 1 |
| chr23_-_33753484 | 1.47 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr11_+_5724343 | 1.45 |
ENSDART00000172911
|
r3hdm4
|
R3H domain containing 4 |
| KN150137v1_+_17956 | 1.39 |
ENSDART00000164745
|
BPHL
|
biphenyl hydrolase like |
| chr5_+_12894381 | 1.39 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr20_+_27398061 | 1.38 |
ENSDART00000153440
|
BX255913.1
|
ENSDARG00000096709 |
| chr12_-_926596 | 1.38 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr15_-_41732606 | 1.35 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr1_+_57891538 | 1.33 |
ENSDART00000158425
|
FO704618.1
|
ENSDARG00000105176 |
| chr10_+_16543577 | 1.32 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr20_-_50211708 | 1.31 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr18_+_20045293 | 1.28 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr24_-_39315552 | 1.28 |
|
|
|
| chr23_+_2847589 | 1.27 |
ENSDART00000156954
|
plcg1
|
phospholipase C, gamma 1 |
| chr23_-_33848571 | 1.25 |
ENSDART00000027959
|
racgap1
|
Rac GTPase activating protein 1 |
| chr15_-_41732430 | 1.24 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr25_+_2252667 | 1.20 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr13_+_35730680 | 1.20 |
ENSDART00000017202
|
kcnk1b
|
potassium channel, subfamily K, member 1b |
| chr21_+_21871674 | 1.19 |
|
|
|
| chr10_-_35488523 | 1.16 |
|
|
|
| chr17_-_25813136 | 1.15 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr19_-_6503643 | 1.14 |
|
|
|
| chr17_+_48041427 | 1.12 |
ENSDART00000160219
|
slc39a9
|
solute carrier family 39, member 9 |
| chr21_+_10689336 | 1.10 |
ENSDART00000146248
|
znf532
|
zinc finger protein 532 |
| chr7_+_20092235 | 1.09 |
ENSDART00000139274
|
ponzr1
|
plac8 onzin related protein 1 |
| chr25_-_28920272 | 1.09 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr16_-_44910578 | 1.09 |
ENSDART00000157375
|
arhgap33
|
Rho GTPase activating protein 33 |
| chr25_+_13695224 | 1.09 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr21_-_2261748 | 1.08 |
ENSDART00000172041
ENSDART00000161554 |
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr2_-_37762426 | 1.08 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr3_+_32401005 | 1.07 |
ENSDART00000150981
|
nosip
|
nitric oxide synthase interacting protein |
| chr15_-_41732304 | 1.06 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr4_+_4825409 | 1.03 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr8_-_47215639 | 1.02 |
ENSDART00000147606
|
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr21_+_22293328 | 0.98 |
ENSDART00000157839
|
nadk2
|
NAD kinase 2, mitochondrial |
| chr5_-_26349966 | 0.98 |
ENSDART00000050542
|
htra4
|
HtrA serine peptidase 4 |
| chr19_+_2834491 | 0.97 |
ENSDART00000112414
|
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr17_+_32412739 | 0.97 |
ENSDART00000156197
|
CR627496.1
|
ENSDARG00000097538 |
| chr9_+_31265296 | 0.94 |
ENSDART00000132478
|
BX470183.1
|
ENSDARG00000095476 |
| chr10_+_39972154 | 0.94 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr3_-_14421675 | 0.94 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr12_+_37039816 | 0.92 |
ENSDART00000162922
|
CR456627.1
|
ENSDARG00000104543 |
| chr18_+_20044862 | 0.91 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr8_+_26391153 | 0.89 |
ENSDART00000053447
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr6_-_16267327 | 0.86 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr10_+_16544000 | 0.84 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr3_-_15861110 | 0.84 |
ENSDART00000080672
|
mrps34
|
mitochondrial ribosomal protein S34 |
| chr10_+_43578519 | 0.83 |
ENSDART00000132486
|
rasa1b
|
RAS p21 protein activator (GTPase activating protein) 1b |
| chr17_+_25163592 | 0.82 |
ENSDART00000100277
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr18_+_20045221 | 0.79 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr4_-_14894468 | 0.77 |
|
|
|
| chr7_+_28773716 | 0.75 |
ENSDART00000086871
|
gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr6_-_1315025 | 0.74 |
|
|
|
| chr15_-_41732512 | 0.72 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr7_+_23881828 | 0.72 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr25_-_12838540 | 0.72 |
ENSDART00000167362
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr3_-_36235222 | 0.70 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr15_-_23786239 | 0.70 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr12_+_2769741 | 0.66 |
ENSDART00000178262
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr8_+_49103201 | 0.66 |
ENSDART00000032355
|
zgc:56525
|
zgc:56525 |
| chr13_+_21543950 | 0.65 |
ENSDART00000078567
ENSDART00000165166 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr4_+_4825628 | 0.64 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr14_+_1106141 | 0.64 |
ENSDART00000106708
|
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr18_+_45648360 | 0.63 |
ENSDART00000010256
|
eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr22_+_25911875 | 0.63 |
ENSDART00000157842
|
dnaja3b
|
DnaJ (Hsp40) homolog, subfamily A, member 3B |
| chr8_+_24721104 | 0.63 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr21_+_2728070 | 0.62 |
|
|
|
| chr23_+_44795356 | 0.60 |
ENSDART00000145905
|
eno3
|
enolase 3, (beta, muscle) |
| chr6_+_37551455 | 0.59 |
ENSDART00000160960
ENSDART00000039934 ENSDART00000136585 |
slc9a7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr2_-_47020452 | 0.58 |
|
|
|
| chr19_+_33506239 | 0.57 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr3_+_32400841 | 0.57 |
ENSDART00000150981
|
nosip
|
nitric oxide synthase interacting protein |
| chr24_-_31155315 | 0.56 |
ENSDART00000168398
|
hccsb
|
holocytochrome c synthase b |
| chr6_-_1315162 | 0.55 |
|
|
|
| chr18_-_5004815 | 0.53 |
|
|
|
| chr2_+_2977250 | 0.53 |
|
|
|
| chr8_-_48858897 | 0.53 |
ENSDART00000122458
ENSDART00000010591 |
wrap73
|
WD repeat containing, antisense to TP73 |
| chr17_+_20549925 | 0.52 |
ENSDART00000113936
|
ENSDARG00000074663
|
ENSDARG00000074663 |
| chr7_-_44631642 | 0.52 |
ENSDART00000124342
|
fam96b
|
family with sequence similarity 96, member B |
| chr15_-_41732567 | 0.51 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr15_+_847111 | 0.50 |
ENSDART00000158679
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr19_-_18672697 | 0.49 |
|
|
|
| chr1_+_57891569 | 0.45 |
ENSDART00000158425
|
FO704618.1
|
ENSDARG00000105176 |
| chr16_+_42383980 | 0.45 |
|
|
|
| chr10_+_45056951 | 0.43 |
ENSDART00000158553
|
ENSDARG00000101143
|
ENSDARG00000101143 |
| chr25_-_35595100 | 0.41 |
|
|
|
| chr25_-_22954929 | 0.41 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr5_-_40542596 | 0.41 |
ENSDART00000134891
|
zfr
|
zinc finger RNA binding protein |
| chr20_-_51494625 | 0.40 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr8_-_410733 | 0.40 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr21_-_11274574 | 0.37 |
ENSDART00000151000
ENSDART00000151465 |
zgc:162472
|
zgc:162472 |
| chr9_-_46609148 | 0.36 |
|
|
|
| chr24_-_39315521 | 0.30 |
|
|
|
| chr4_+_75469488 | 0.30 |
ENSDART00000144892
|
znf1009
|
zinc finger protein 1009 |
| chr23_-_33753634 | 0.30 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr14_+_21725253 | 0.29 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr16_+_10950751 | 0.22 |
ENSDART00000128762
|
dedd1
|
death effector domain-containing 1 |
| chr19_+_4128351 | 0.13 |
ENSDART00000161676
|
bty
|
bloodthirsty |
| chr8_-_47215701 | 0.12 |
ENSDART00000060883
|
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr3_+_35478909 | 0.11 |
ENSDART00000132703
|
traf7
|
TNF receptor-associated factor 7 |
| chr7_-_32859341 | 0.10 |
ENSDART00000127006
ENSDART00000143768 |
mns1
|
meiosis-specific nuclear structural 1 |
| chr3_-_40020899 | 0.10 |
ENSDART00000025285
|
drg2
|
developmentally regulated GTP binding protein 2 |
| chr13_+_37527958 | 0.08 |
ENSDART00000141988
|
phf3
|
PHD finger protein 3 |
| chr18_+_2902170 | 0.07 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr6_-_1315096 | 0.06 |
|
|
|
| chr8_+_31325461 | 0.06 |
|
|
|
| chr17_+_12504341 | 0.06 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr24_-_21740552 | 0.06 |
|
|
|
| chr16_+_10950160 | 0.04 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr6_-_16267239 | 0.03 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr20_+_41123859 | 0.03 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr5_+_23651005 | 0.02 |
ENSDART00000089851
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.8 | 2.5 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.8 | 3.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 3.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 2.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 7.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.4 | 1.3 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.4 | 2.3 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.3 | 3.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 3.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.3 | 2.4 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.2 | 0.7 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 1.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.7 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.2 | 3.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 4.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 0.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 1.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 8.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 3.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 12.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 6.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0048680 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 1.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 2.0 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 7.3 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 1.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 4.9 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.8 | 5.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 1.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 3.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.3 | 0.9 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 3.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 0.7 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 9.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 3.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.8 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.4 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 2.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 4.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.6 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.6 | 4.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 1.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.3 | 1.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.3 | 0.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.3 | 9.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 3.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 10.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 3.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 8.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.8 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 3.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.0 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 3.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 8.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 2.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 1.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 1.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 4.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.6 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 2.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 7.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |