Project
DANIO-CODE
Navigation
Downloads
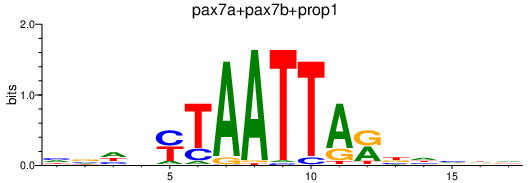
Results for pax7a+pax7b+prop1
Z-value: 0.78
Transcription factors associated with pax7a+pax7b+prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prop1
|
ENSDARG00000039756 | PROP paired-like homeobox 1 |
|
pax7b
|
ENSDARG00000070818 | paired box 7b |
|
pax7a
|
ENSDARG00000100398 | paired box 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax7b | dr10_dc_chr23_+_20936600_20936609 | -0.96 | 4.3e-09 | Click! |
| pax7a | dr10_dc_chr11_+_40652239_40652375 | -0.94 | 1.0e-07 | Click! |
Activity profile of pax7a+pax7b+prop1 motif
Sorted Z-values of pax7a+pax7b+prop1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax7a+pax7b+prop1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37303599 | 2.23 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr10_-_34058331 | 2.14 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr10_+_33449922 | 2.06 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr10_-_25246786 | 1.99 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr24_+_19270877 | 1.99 |
|
|
|
| chr22_-_20670164 | 1.92 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr8_+_45326435 | 1.89 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_+_6289363 | 1.88 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr16_-_24727689 | 1.74 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr11_+_12754166 | 1.70 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr16_+_47283253 | 1.70 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr2_+_6341404 | 1.69 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr1_-_54391298 | 1.68 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr3_+_17562348 | 1.62 |
|
|
|
| chr10_+_17277353 | 1.59 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr21_-_32027717 | 1.58 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr9_+_8412489 | 1.56 |
ENSDART00000144851
|
zgc:113984
|
zgc:113984 |
| chr25_+_5845303 | 1.48 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr4_+_4825461 | 1.46 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr21_+_22598805 | 1.43 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr14_+_22159893 | 1.41 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr6_-_50731449 | 1.38 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr4_-_62022037 | 1.37 |
|
|
|
| chr19_+_2942485 | 1.36 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr25_+_7067115 | 1.33 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr14_-_14353451 | 1.25 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr25_+_7067043 | 1.23 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr25_-_13394261 | 1.22 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr15_+_35076414 | 1.21 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr6_-_8156471 | 1.20 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr9_+_23813071 | 1.20 |
|
|
|
| chr23_+_19663746 | 1.19 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr15_-_16241412 | 1.16 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_-_53165501 | 1.16 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr16_+_47283374 | 1.15 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr22_+_2735606 | 1.14 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr15_+_40340914 | 1.13 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr20_+_54404987 | 1.13 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr11_+_17849608 | 1.12 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr25_+_19302983 | 1.09 |
|
|
|
| chr12_-_33256934 | 1.08 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr20_-_29961498 | 1.07 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr14_-_49777408 | 1.07 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr8_+_11287550 | 1.05 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr20_-_29961589 | 1.03 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr6_+_18941289 | 1.01 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr12_-_33256754 | 0.99 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_-_33256671 | 0.98 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_-_5186692 | 0.97 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_-_26675055 | 0.96 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_-_11162837 | 0.96 |
|
|
|
| chr6_+_18941186 | 0.95 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr22_+_1837102 | 0.95 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr6_+_40925259 | 0.95 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_+_8314513 | 0.94 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| KN150663v1_-_3381 | 0.94 |
|
|
|
| chr11_+_6012910 | 0.92 |
ENSDART00000161001
|
gtpbp3
|
GTP binding protein 3 |
| chr22_+_11943005 | 0.92 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr13_-_51743281 | 0.91 |
|
|
|
| chr6_-_10676857 | 0.88 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr11_-_44756789 | 0.88 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr15_-_25025251 | 0.87 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr23_-_19299279 | 0.85 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr4_+_13902603 | 0.83 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr2_+_8314372 | 0.83 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr19_-_1571878 | 0.81 |
|
|
|
| chr10_-_17214368 | 0.80 |
|
|
|
| chr1_+_35253862 | 0.79 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr19_-_18664670 | 0.79 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr24_-_2454189 | 0.78 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr2_-_57826189 | 0.78 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr12_-_33257026 | 0.77 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_27477153 | 0.77 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr21_+_21871674 | 0.76 |
|
|
|
| chr14_+_45685886 | 0.75 |
|
|
|
| chr12_+_22459218 | 0.74 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr10_+_33450122 | 0.74 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr21_-_13026036 | 0.74 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr9_-_34700736 | 0.73 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr18_+_19467527 | 0.73 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr12_+_33257120 | 0.73 |
|
|
|
| chr9_-_32347822 | 0.73 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr4_+_4825628 | 0.73 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr6_+_18941135 | 0.73 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr23_-_33753484 | 0.73 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr12_-_33256599 | 0.73 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_+_46362661 | 0.72 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr8_+_11387135 | 0.72 |
|
|
|
| chr1_+_13244145 | 0.72 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr5_+_1109967 | 0.72 |
ENSDART00000046781
ENSDART00000163101 |
rnf185
|
ring finger protein 185 |
| chr5_+_6005049 | 0.71 |
|
|
|
| chr18_+_15695287 | 0.71 |
|
|
|
| chr10_+_43953171 | 0.71 |
|
|
|
| chr20_-_46079529 | 0.70 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr16_-_31386496 | 0.69 |
|
|
|
| chr20_+_29306863 | 0.69 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_-_43725091 | 0.68 |
|
|
|
| chr17_-_21029377 | 0.68 |
ENSDART00000155300
|
bicc1a
|
BicC family RNA binding protein 1a |
| chr19_+_1743359 | 0.67 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr15_-_43402935 | 0.66 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_+_27100350 | 0.66 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr17_-_9794176 | 0.65 |
|
|
|
| chr8_-_44247277 | 0.64 |
|
|
|
| chr14_+_47190202 | 0.64 |
ENSDART00000074116
|
rab33ba
|
RAB33B, member RAS oncogene family a |
| chr5_-_3673698 | 0.64 |
|
|
|
| chr24_-_21788942 | 0.63 |
ENSDART00000113092
|
abhd10b
|
abhydrolase domain containing 10b |
| chr20_+_25687135 | 0.62 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr17_+_12788541 | 0.62 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr15_+_14656886 | 0.62 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr25_-_19927083 | 0.62 |
ENSDART00000140182
|
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr22_-_10511906 | 0.62 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr21_+_31113731 | 0.61 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr12_+_22459177 | 0.61 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_2942373 | 0.61 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr8_-_53593850 | 0.60 |
|
|
|
| chr14_-_6901783 | 0.60 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr23_+_26099718 | 0.60 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr17_+_32579392 | 0.60 |
ENSDART00000112559
|
gcfc2
|
GC-rich sequence DNA-binding factor 2 |
| chr15_-_16241500 | 0.60 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr24_-_34794538 | 0.60 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr10_-_43874597 | 0.59 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| KN150200v1_+_7856 | 0.59 |
|
|
|
| chr7_+_22042296 | 0.59 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr25_-_19926991 | 0.59 |
ENSDART00000140182
|
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr7_-_73204361 | 0.58 |
ENSDART00000157500
|
AL929105.1
|
ENSDARG00000105194 |
| chr10_-_25808063 | 0.58 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| KN150200v1_+_7782 | 0.58 |
|
|
|
| chr2_-_8002471 | 0.58 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr2_+_8314844 | 0.58 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr17_-_22553428 | 0.58 |
ENSDART00000079390
|
exo1
|
exonuclease 1 |
| chr10_+_1801745 | 0.58 |
|
|
|
| chr20_+_46588659 | 0.57 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr17_-_25630635 | 0.57 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr10_-_21971084 | 0.57 |
ENSDART00000174954
|
CABZ01031892.1
|
ENSDARG00000107280 |
| chr7_-_9628384 | 0.57 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr22_+_17803347 | 0.57 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr17_+_49540985 | 0.57 |
|
|
|
| chr20_-_46079578 | 0.56 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr21_-_2183869 | 0.56 |
ENSDART00000163659
|
zgc:162971
|
zgc:162971 |
| chr21_-_25765319 | 0.56 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr15_+_2556689 | 0.56 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr16_+_12726556 | 0.56 |
|
|
|
| chr8_-_16638825 | 0.56 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr2_+_6341345 | 0.55 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_+_29306677 | 0.55 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr22_+_10668889 | 0.54 |
ENSDART00000081228
|
abhd14a
|
abhydrolase domain containing 14A |
| chr19_+_10742344 | 0.53 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr14_-_26127173 | 0.53 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr20_-_55094208 | 0.53 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr3_-_15517844 | 0.53 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr12_+_10405904 | 0.52 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr1_-_17544731 | 0.52 |
ENSDART00000143157
|
AL929210.1
|
ENSDARG00000094943 |
| chr7_+_23752492 | 0.52 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_37303707 | 0.51 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr20_-_33272243 | 0.51 |
ENSDART00000178752
ENSDART00000047834 |
nbas
|
neuroblastoma amplified sequence |
| chr23_-_15234716 | 0.51 |
ENSDART00000158943
|
ndrg3b
|
ndrg family member 3b |
| chr19_+_31280451 | 0.51 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr16_-_28723759 | 0.51 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr22_-_37861903 | 0.50 |
ENSDART00000085931
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr3_-_15517767 | 0.50 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr8_+_30103357 | 0.50 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr22_+_1335355 | 0.50 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr1_+_8010026 | 0.50 |
ENSDART00000152142
|
zgc:77849
|
zgc:77849 |
| chr8_-_49739908 | 0.50 |
ENSDART00000138810
|
gkap1
|
G kinase anchoring protein 1 |
| chr7_+_24374240 | 0.49 |
ENSDART00000087691
|
gba2
|
glucosidase, beta (bile acid) 2 |
| chr22_+_35229352 | 0.49 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr4_-_2370333 | 0.49 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr6_+_56224631 | 0.49 |
ENSDART00000174612
ENSDART00000177824 |
FO834918.1
|
ENSDARG00000106015 |
| chr13_+_40347099 | 0.49 |
|
|
|
| chr21_-_39132507 | 0.48 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr9_+_8412584 | 0.48 |
ENSDART00000133880
ENSDART00000142233 |
zgc:113984
|
zgc:113984 |
| chr17_-_21029294 | 0.48 |
ENSDART00000155300
|
bicc1a
|
BicC family RNA binding protein 1a |
| chr6_-_9440411 | 0.48 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr4_-_20119812 | 0.48 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr15_-_25025058 | 0.48 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr24_-_17261929 | 0.48 |
ENSDART00000153858
|
BX005392.2
|
ENSDARG00000096996 |
| chr16_+_9835124 | 0.47 |
ENSDART00000020859
|
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr23_+_43968576 | 0.47 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr21_-_4913068 | 0.47 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr11_-_12744413 | 0.46 |
ENSDART00000104143
|
txlng
|
taxilin gamma |
| KN149830v1_-_31870 | 0.46 |
ENSDART00000167653
|
zgc:194336
|
zgc:194336 |
| chr23_-_29467861 | 0.46 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr20_+_24047842 | 0.45 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr3_+_52290252 | 0.45 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr3_-_34208507 | 0.45 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr15_-_47260834 | 0.45 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr2_+_11335970 | 0.45 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr17_-_8155740 | 0.44 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr16_-_31664756 | 0.44 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr5_-_68848113 | 0.44 |
ENSDART00000097249
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr19_+_5562075 | 0.44 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr12_+_22459371 | 0.44 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr8_-_22305457 | 0.43 |
ENSDART00000149517
|
cep104
|
centrosomal protein 104 |
| chr8_+_21127033 | 0.43 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr22_-_35218207 | 0.43 |
ENSDART00000104687
|
pfn2
|
profilin 2 |
| chr11_+_30006715 | 0.43 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr20_+_54404819 | 0.43 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr9_+_2523927 | 0.43 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr11_-_2437361 | 0.42 |
|
|
|
| chr10_-_43874646 | 0.42 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.5 | 1.4 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.4 | 1.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 1.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 0.8 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 0.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 1.4 | GO:0030237 | female sex determination(GO:0030237) |
| 0.2 | 0.7 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.2 | 2.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.8 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.2 | 0.9 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.5 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 2.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 2.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.2 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0051977 | lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.1 | 0.6 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 4.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0003128 | heart field specification(GO:0003128) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.5 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.2 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.1 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0022406 | membrane docking(GO:0022406) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 2.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 0.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.4 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 2.1 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.5 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.4 | 1.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.4 | 2.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 1.0 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 1.7 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.3 | 1.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 2.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 2.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 1.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.6 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 2.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.1 | 2.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0015562 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 3.2 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.5 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 2.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.3 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 3.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 4.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |