Project
DANIO-CODE
Navigation
Downloads
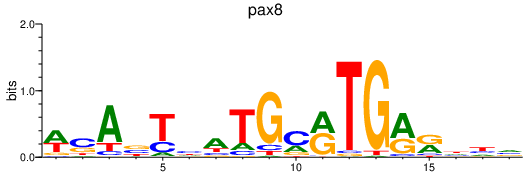
Results for pax8
Z-value: 0.27
Transcription factors associated with pax8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax8
|
ENSDARG00000015879 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax8 | dr10_dc_chr5_-_71643185_71643209 | -0.59 | 1.7e-02 | Click! |
Activity profile of pax8 motif
Sorted Z-values of pax8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_25297360 | 0.46 |
ENSDART00000138215
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr16_-_34240818 | 0.40 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr4_+_1727494 | 0.34 |
ENSDART00000149448
|
AL929171.1
|
ENSDARG00000095920 |
| chr9_+_35205519 | 0.32 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr5_-_36741099 | 0.29 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr9_-_23954640 | 0.26 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr5_-_36741219 | 0.26 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr5_-_36741034 | 0.25 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr23_-_24772871 | 0.25 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr16_-_43107682 | 0.24 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr10_+_29883772 | 0.24 |
ENSDART00000100032
|
hyou1
|
hypoxia up-regulated 1 |
| chr12_-_4212149 | 0.24 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr19_-_2084484 | 0.22 |
|
|
|
| chr24_-_25297298 | 0.21 |
ENSDART00000138215
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr12_+_19226572 | 0.21 |
ENSDART00000066388
|
tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr10_+_29884057 | 0.20 |
ENSDART00000100032
|
hyou1
|
hypoxia up-regulated 1 |
| chr7_-_29084775 | 0.20 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr7_+_19300487 | 0.19 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr9_-_22014861 | 0.19 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr4_+_56996802 | 0.19 |
|
|
|
| chr9_+_2538387 | 0.19 |
|
|
|
| chr9_-_23954761 | 0.18 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr13_+_32323781 | 0.18 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a (all-trans/9-cis/11-cis) |
| chr9_-_32532691 | 0.17 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr21_-_20083822 | 0.17 |
ENSDART00000065664
|
dusp4
|
dual specificity phosphatase 4 |
| chr11_-_40239946 | 0.17 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr13_-_35719176 | 0.16 |
ENSDART00000171371
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr16_+_46728897 | 0.14 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr9_-_23954690 | 0.14 |
ENSDART00000027212
|
ENSDARG00000006848
|
ENSDARG00000006848 |
| chr4_+_90885 | 0.14 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_-_50588444 | 0.13 |
ENSDART00000151500
|
mtss1
|
metastasis suppressor 1 |
| chr9_+_35205252 | 0.13 |
ENSDART00000100701
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr22_+_3994627 | 0.13 |
ENSDART00000166768
|
timm44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr20_+_15264912 | 0.12 |
ENSDART00000063877
|
prdx6
|
peroxiredoxin 6 |
| chr12_-_4212095 | 0.12 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr11_-_25224150 | 0.11 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr11_+_18856937 | 0.11 |
|
|
|
| chr12_+_23300259 | 0.11 |
|
|
|
| chr8_-_7278846 | 0.11 |
ENSDART00000162228
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr16_+_50355723 | 0.11 |
ENSDART00000166664
|
ENSDARG00000102916
|
ENSDARG00000102916 |
| chr12_+_19226616 | 0.10 |
ENSDART00000066388
|
tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr22_-_16128314 | 0.10 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr9_+_23954807 | 0.10 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr10_+_24690784 | 0.10 |
ENSDART00000147974
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr24_+_31396974 | 0.09 |
ENSDART00000168837
|
fam168b
|
family with sequence similarity 168, member B |
| chr19_-_2084281 | 0.08 |
|
|
|
| chr20_-_14784875 | 0.08 |
ENSDART00000063857
|
scrn2
|
secernin 2 |
| chr9_+_23954669 | 0.08 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr15_+_16451221 | 0.08 |
ENSDART00000101157
|
flot2b
|
flotillin 2b |
| chr9_-_32532616 | 0.07 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr3_-_15960288 | 0.06 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr17_+_612173 | 0.06 |
|
|
|
| chr23_-_24772748 | 0.05 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr22_+_18904372 | 0.04 |
ENSDART00000131131
|
bsg
|
basigin |
| chr9_+_23954607 | 0.04 |
ENSDART00000145120
|
ENSDARG00000040445
|
ENSDARG00000040445 |
| chr5_+_68055581 | 0.04 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr9_-_22014768 | 0.04 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr6_-_47841704 | 0.04 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr23_-_24772650 | 0.03 |
ENSDART00000104031
|
sdf4
|
stromal cell derived factor 4 |
| chr25_+_18378608 | 0.03 |
ENSDART00000158142
ENSDART00000073563 |
tes
|
testis derived transcript (3 LIM domains) |
| chr5_+_68055760 | 0.02 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr10_+_24690680 | 0.02 |
ENSDART00000147974
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr24_-_21828216 | 0.02 |
ENSDART00000030592
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr13_-_35719075 | 0.02 |
ENSDART00000171371
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| KN149962v1_-_109672 | 0.02 |
|
|
|
| chr23_-_7191380 | 0.02 |
ENSDART00000176914
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr7_-_28278230 | 0.01 |
ENSDART00000054368
|
st5
|
suppression of tumorigenicity 5 |
| chr7_-_51210658 | 0.01 |
ENSDART00000174026
|
nhsl2
|
NHS-like 2 |
| chr10_+_36094175 | 0.01 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr20_+_32321484 | 0.01 |
ENSDART00000147448
|
sesn1
|
sestrin 1 |
| chr18_+_27242463 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.2 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0048855 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |