Project
DANIO-CODE
Navigation
Downloads
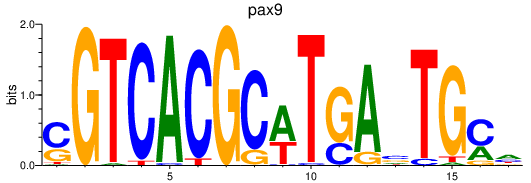
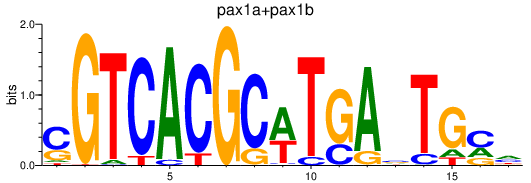
Results for pax9_pax1a+pax1b
Z-value: 0.86
Transcription factors associated with pax9_pax1a+pax1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax9
|
ENSDARG00000053829 | paired box 9 |
|
pax1a
|
ENSDARG00000008203 | paired box 1a |
|
pax1b
|
ENSDARG00000073814 | paired box 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax9 | dr10_dc_chr17_-_38296557_38296571 | 0.65 | 6.7e-03 | Click! |
Activity profile of pax9_pax1a+pax1b motif
Sorted Z-values of pax9_pax1a+pax1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pax9_pax1a+pax1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_41790437 | 4.44 |
ENSDART00000159135
ENSDART00000158456 ENSDART00000160069 |
lama1
|
laminin, alpha 1 |
| chr18_+_48426375 | 3.26 |
|
|
|
| chr21_+_13764828 | 2.81 |
ENSDART00000171306
|
stxbp1a
|
syntaxin binding protein 1a |
| chr15_+_41878670 | 2.58 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr2_-_44429891 | 2.46 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr23_+_157661 | 2.18 |
|
|
|
| chr5_+_296856 | 1.83 |
|
|
|
| chr21_+_13764917 | 1.76 |
ENSDART00000015629
|
stxbp1a
|
syntaxin binding protein 1a |
| chr4_-_16461748 | 1.71 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr12_+_10594531 | 1.68 |
|
|
|
| chr18_-_3244618 | 1.61 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr16_-_9108885 | 1.54 |
ENSDART00000153785
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr1_+_38834751 | 1.32 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr17_+_1610578 | 1.24 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr12_-_7820226 | 1.21 |
ENSDART00000149594
ENSDART00000148866 |
ank3b
|
ankyrin 3b |
| chr17_+_794430 | 1.06 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr20_+_29685096 | 0.92 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr5_+_52198613 | 0.80 |
ENSDART00000162459
|
scarb2a
|
scavenger receptor class B, member 2a |
| chr10_+_39211384 | 0.76 |
|
|
|
| chr9_-_24219908 | 0.73 |
ENSDART00000144163
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr16_+_20932876 | 0.71 |
ENSDART00000079343
|
jazf1b
|
JAZF zinc finger 1b |
| chr13_-_24248675 | 0.68 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr14_+_23220869 | 0.67 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr10_-_39211281 | 0.66 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr5_-_35143549 | 0.65 |
ENSDART00000157881
|
ophn1
|
oligophrenin 1 |
| chr3_-_49242655 | 0.65 |
ENSDART00000142200
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr24_-_36840203 | 0.44 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr6_-_26897413 | 0.31 |
|
|
|
| chr8_-_16614882 | 0.30 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr20_+_29685331 | 0.30 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr9_-_23177842 | 0.19 |
|
|
|
| chr23_+_46015863 | 0.16 |
ENSDART00000012234
|
syap1
|
synapse associated protein 1 |
| chr8_+_48976926 | 0.05 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr24_+_3447129 | 0.02 |
ENSDART00000134598
|
wdr37
|
WD repeat domain 37 |
| chr20_+_12809388 | 0.01 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr7_+_26274252 | 0.00 |
ENSDART00000164824
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.7 | 4.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 1.3 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.7 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 4.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.7 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 4.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |