Project
DANIO-CODE
Navigation
Downloads
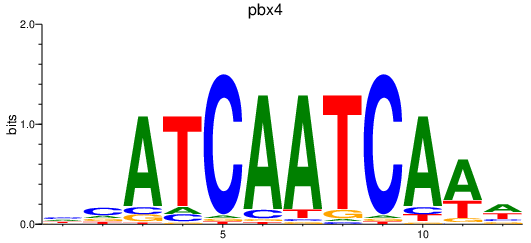
Results for pbx4
Z-value: 1.14
Transcription factors associated with pbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pbx4
|
ENSDARG00000052150 | pre-B-cell leukemia transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pbx4 | dr10_dc_chr3_+_52745606_52745690 | -0.85 | 2.5e-05 | Click! |
Activity profile of pbx4 motif
Sorted Z-values of pbx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pbx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_25426346 | 8.35 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr21_-_23271127 | 5.00 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr13_-_5440923 | 4.63 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr19_+_25009921 | 3.59 |
|
|
|
| chr17_+_52736192 | 3.37 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr24_-_6868340 | 2.72 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr23_+_32573474 | 2.37 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr6_-_14821305 | 2.25 |
|
|
|
| chr1_+_25378105 | 2.20 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr25_+_24520476 | 2.17 |
|
|
|
| chr19_-_5429428 | 2.15 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr24_+_18804086 | 2.10 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr20_-_42805703 | 2.08 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr19_+_20209561 | 2.07 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr5_+_34857986 | 2.03 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr7_+_26357875 | 1.97 |
ENSDART00000101044
|
hsbp1a
|
heat shock factor binding protein 1a |
| chr3_-_32202533 | 1.94 |
ENSDART00000155757
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr16_-_44682957 | 1.87 |
|
|
|
| chr1_-_46292847 | 1.85 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_+_2391223 | 1.84 |
|
|
|
| chr1_-_9387290 | 1.78 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr17_-_52735250 | 1.77 |
|
|
|
| chr6_+_29800606 | 1.74 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr8_-_25055333 | 1.69 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr21_-_43083936 | 1.67 |
ENSDART00000040169
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr8_+_49789789 | 1.67 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr7_+_16610457 | 1.66 |
|
|
|
| chr4_+_13811932 | 1.60 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr12_+_48643983 | 1.58 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr9_-_13383818 | 1.58 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr13_-_24316940 | 1.57 |
ENSDART00000127741
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr3_-_45779817 | 1.57 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr23_+_20936419 | 1.49 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr12_-_26760324 | 1.48 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr17_-_52735615 | 1.47 |
|
|
|
| chr22_-_5652624 | 1.46 |
ENSDART00000127688
ENSDART00000012686 |
dnase1l4.1
|
deoxyribonuclease 1 like 4, tandem duplicate 1 |
| chr24_-_6048914 | 1.42 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr5_-_20310793 | 1.38 |
ENSDART00000147639
|
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr13_-_5440751 | 1.36 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr6_+_4712785 | 1.36 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr16_-_16433191 | 1.35 |
ENSDART00000022685
|
nradd
|
neurotrophin receptor associated death domain |
| chr9_-_23406315 | 1.34 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr21_-_17259886 | 1.34 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr10_+_2779947 | 1.34 |
ENSDART00000131435
|
aebp1
|
AE binding protein 1 |
| chr17_+_23892232 | 1.34 |
|
|
|
| chr13_+_16130123 | 1.33 |
ENSDART00000170855
|
mocos
|
molybdenum cofactor sulfurase |
| chr16_-_54638972 | 1.33 |
|
|
|
| chr4_-_13996902 | 1.31 |
ENSDART00000147955
|
prickle1b
|
prickle homolog 1b |
| chr18_+_48426375 | 1.30 |
|
|
|
| chr2_-_21960233 | 1.29 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr5_+_49103778 | 1.29 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr23_-_15381030 | 1.29 |
|
|
|
| chr23_+_20936374 | 1.26 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr8_+_47909655 | 1.24 |
|
|
|
| chr13_-_31165867 | 1.24 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr5_+_296856 | 1.23 |
|
|
|
| chr2_-_10394639 | 1.20 |
ENSDART00000128535
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr23_-_15442032 | 1.20 |
ENSDART00000082060
|
ENSDARG00000078145
|
ENSDARG00000078145 |
| chr3_-_15918262 | 1.20 |
ENSDART00000157315
|
ENSDARG00000097322
|
ENSDARG00000097322 |
| chr13_-_5440558 | 1.17 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr3_-_37558102 | 1.17 |
ENSDART00000151810
|
gpatch8
|
G patch domain containing 8 |
| chr10_-_44509732 | 1.16 |
|
|
|
| chr9_-_33916709 | 1.16 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr8_+_5222065 | 1.15 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr10_+_44509370 | 1.14 |
|
|
|
| chr7_-_18346474 | 1.14 |
|
|
|
| chr12_+_2835894 | 1.14 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr9_+_32267615 | 1.11 |
|
|
|
| chr1_-_25377787 | 1.11 |
|
|
|
| chr10_-_26782374 | 1.11 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr20_-_29148810 | 1.10 |
ENSDART00000140350
|
thbs1b
|
thrombospondin 1b |
| chr9_-_2588401 | 1.10 |
ENSDART00000161018
|
sp9
|
sp9 transcription factor |
| chr8_+_30491476 | 1.09 |
ENSDART00000062073
|
ENSDARG00000042329
|
ENSDARG00000042329 |
| chr21_+_22386643 | 1.09 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr16_-_19154920 | 1.09 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr6_+_30441419 | 1.08 |
|
|
|
| chr20_+_15119519 | 1.07 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr11_+_43559400 | 1.07 |
|
|
|
| chr10_+_45499009 | 1.04 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr10_+_9134634 | 1.03 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr2_-_1752776 | 1.03 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr16_+_21121428 | 1.02 |
|
|
|
| chr10_-_35293024 | 1.01 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr5_+_44407763 | 1.01 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr19_-_7576069 | 1.00 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr23_+_24711233 | 0.97 |
|
|
|
| chr22_-_11408859 | 0.97 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr18_+_5707066 | 0.96 |
|
|
|
| chr7_+_56375651 | 0.96 |
ENSDART00000112242
|
zgc:194679
|
zgc:194679 |
| chr20_+_15119754 | 0.96 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr3_-_47328771 | 0.95 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr16_+_46902372 | 0.95 |
ENSDART00000177679
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr13_-_31304614 | 0.95 |
|
|
|
| chr15_+_32410860 | 0.93 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr1_-_7744605 | 0.92 |
ENSDART00000033917
|
syngr3b
|
synaptogyrin 3b |
| chr4_-_16344954 | 0.90 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr24_+_35899507 | 0.89 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr6_-_2294751 | 0.89 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr17_+_52736535 | 0.89 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr17_-_19606453 | 0.86 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr12_-_11419332 | 0.86 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr17_+_23102259 | 0.86 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_+_19737802 | 0.85 |
ENSDART00000153643
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr3_-_16077831 | 0.84 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr16_-_16244210 | 0.83 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr16_-_50024848 | 0.83 |
ENSDART00000161782
|
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr6_-_50705420 | 0.82 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr22_+_528076 | 0.82 |
|
|
|
| chr3_+_23621843 | 0.82 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr19_-_27284726 | 0.80 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr21_-_37914590 | 0.80 |
|
|
|
| chr18_-_25002972 | 0.79 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_+_48708504 | 0.78 |
ENSDART00000008468
|
msx1b
|
muscle segment homeobox 1b |
| chr12_-_48643687 | 0.78 |
ENSDART00000178135
ENSDART00000176040 |
CABZ01078412.1
|
ENSDARG00000107260 |
| chr2_-_1753058 | 0.78 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr7_+_42124857 | 0.77 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr20_+_48971948 | 0.76 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr3_-_44113070 | 0.75 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr4_-_20764499 | 0.74 |
ENSDART00000023337
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr8_+_8056616 | 0.74 |
|
|
|
| chr15_+_3296905 | 0.73 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr5_-_33156615 | 0.73 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr10_-_27780050 | 0.73 |
ENSDART00000138149
|
si:dkey-33o22.1
|
si:dkey-33o22.1 |
| chr21_-_44109455 | 0.72 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr18_-_45892 | 0.72 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr22_+_35090105 | 0.72 |
|
|
|
| chr20_+_18803167 | 0.72 |
ENSDART00000019476
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr21_+_26690161 | 0.71 |
ENSDART00000065392
|
calm3b
|
calmodulin 3b (phosphorylase kinase, delta) |
| chr17_+_52736844 | 0.71 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr19_+_27133834 | 0.71 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr13_+_28286826 | 0.70 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr5_-_24365811 | 0.70 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr2_-_24947660 | 0.70 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr21_-_36886505 | 0.70 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr4_+_9535985 | 0.69 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr1_-_56233548 | 0.68 |
ENSDART00000152597
|
si:ch211-1f22.1
|
si:ch211-1f22.1 |
| chr15_+_895792 | 0.67 |
|
|
|
| chr7_+_15619409 | 0.66 |
ENSDART00000144765
ENSDART00000145946 |
pax6b
|
paired box 6b |
| chr24_+_34227202 | 0.66 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr14_-_205130 | 0.65 |
ENSDART00000035581
ENSDART00000158725 ENSDART00000163549 |
OTOP1
|
otopetrin 1 |
| chr1_-_20132900 | 0.65 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr22_+_35113233 | 0.64 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr8_+_50994889 | 0.63 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr10_-_17383380 | 0.63 |
|
|
|
| chr3_+_22447812 | 0.60 |
ENSDART00000112270
|
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr2_-_7560202 | 0.60 |
ENSDART00000164494
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr6_+_40664212 | 0.59 |
ENSDART00000103842
|
eno1b
|
enolase 1b, (alpha) |
| chr3_+_23621561 | 0.58 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr9_+_32267297 | 0.57 |
|
|
|
| chr20_+_33889082 | 0.57 |
ENSDART00000175023
|
olfml2ba
|
olfactomedin-like 2Ba |
| chr9_+_56507484 | 0.56 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr3_-_15556928 | 0.55 |
ENSDART00000134729
|
BX936308.1
|
ENSDARG00000081299 |
| chr1_-_39509011 | 0.55 |
ENSDART00000146680
|
ENSDARG00000078251
|
ENSDARG00000078251 |
| chr7_+_15623685 | 0.53 |
ENSDART00000146704
|
pax6b
|
paired box 6b |
| chr13_+_50062513 | 0.53 |
ENSDART00000124142
ENSDART00000099537 |
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr16_+_54001592 | 0.52 |
|
|
|
| chr11_-_16067646 | 0.52 |
|
|
|
| chr5_-_19691538 | 0.52 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr2_-_19181629 | 0.51 |
ENSDART00000165782
|
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr5_-_24365560 | 0.51 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr16_+_35917621 | 0.50 |
|
|
|
| chr10_-_17383466 | 0.47 |
|
|
|
| chr4_-_16344835 | 0.47 |
ENSDART00000134449
|
epyc
|
epiphycan |
| chr5_-_48377307 | 0.47 |
ENSDART00000097452
|
si:dkey-172m14.1
|
si:dkey-172m14.1 |
| chr18_+_20836320 | 0.47 |
ENSDART00000090100
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr19_+_14197118 | 0.47 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr19_-_24683045 | 0.45 |
ENSDART00000104083
|
s100v2
|
S100 calcium binding protein V2 |
| chr12_+_34726748 | 0.45 |
ENSDART00000105533
|
enthd2
|
ENTH domain containing 2 |
| chr20_-_54591757 | 0.45 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr12_+_47029281 | 0.44 |
|
|
|
| chr16_-_36090863 | 0.44 |
|
|
|
| chr14_-_1342450 | 0.44 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr15_-_42081757 | 0.44 |
ENSDART00000004338
|
epha4l
|
eph receptor A4, like |
| chr2_-_54028761 | 0.44 |
ENSDART00000178289
|
CABZ01050256.1
|
ENSDARG00000109075 |
| chr17_-_28084988 | 0.43 |
ENSDART00000149654
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr23_+_20936600 | 0.43 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr2_-_52792076 | 0.42 |
ENSDART00000097716
|
zgc:136336
|
zgc:136336 |
| chr22_+_17409030 | 0.41 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr19_+_43524566 | 0.40 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr7_-_20201693 | 0.40 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr7_-_20201358 | 0.39 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr9_-_13383708 | 0.39 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr5_-_24365703 | 0.38 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr8_-_12253026 | 0.38 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr3_-_39117801 | 0.38 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| KN150015v1_+_1935 | 0.38 |
ENSDART00000169907
ENSDART00000160155 |
PDCL3
|
phosducin like 3 |
| chr2_+_34155114 | 0.37 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr13_-_49749469 | 0.36 |
ENSDART00000136165
|
lyst
|
lysosomal trafficking regulator |
| chr20_+_4750766 | 0.35 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr11_+_23012136 | 0.35 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr10_-_14587464 | 0.35 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr5_-_23715514 | 0.34 |
ENSDART00000011878
|
eif4a1b
|
eukaryotic translation initiation factor 4A1B |
| chr16_+_24047857 | 0.33 |
|
|
|
| chr11_-_29569229 | 0.33 |
ENSDART00000069132
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr16_-_19154824 | 0.31 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr23_+_2391317 | 0.31 |
|
|
|
| chr24_+_20391758 | 0.31 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr23_-_18454945 | 0.30 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr15_+_14925920 | 0.29 |
|
|
|
| chr6_-_2294717 | 0.29 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 1.2 | 5.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.8 | 3.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.7 | 2.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.7 | 2.0 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.6 | 1.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.5 | 1.5 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.4 | 7.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.4 | 2.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 2.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.3 | 1.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.2 | 0.7 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.2 | 0.7 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) smooth muscle tissue development(GO:0048745) |
| 0.2 | 0.7 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 1.2 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 0.7 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 1.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 0.6 | GO:0050886 | negative regulation of lipid transport(GO:0032369) endocrine process(GO:0050886) positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 2.0 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 1.2 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 1.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 2.0 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.5 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.1 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 1.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 1.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.8 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 5.7 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 1.6 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.0 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0035143 | embryonic caudal fin morphogenesis(GO:0035124) caudal fin morphogenesis(GO:0035143) |
| 0.0 | 0.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 1.0 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.7 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.2 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.9 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.6 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0030426 | filopodium(GO:0030175) growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.6 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 1.4 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.4 | 1.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 1.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.3 | 2.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 1.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 8.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 12.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 5.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 3.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 3.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 3.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |