Project
DANIO-CODE
Navigation
Downloads
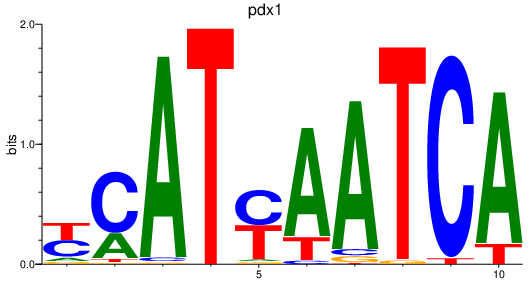
Results for pdx1
Z-value: 1.12
Transcription factors associated with pdx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pdx1
|
ENSDARG00000002779 | pancreatic and duodenal homeobox 1 |
Activity profile of pdx1 motif
Sorted Z-values of pdx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pdx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_25426346 | 8.62 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr21_-_23271127 | 4.44 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr3_-_32686790 | 4.39 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_+_24057260 | 3.78 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr8_-_16662185 | 3.57 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr1_-_19152361 | 3.52 |
ENSDART00000132958
|
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr13_-_5440923 | 3.19 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr19_+_25009921 | 3.02 |
|
|
|
| chr20_-_29517770 | 2.66 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr16_+_24063849 | 2.49 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr12_+_28739504 | 2.36 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr5_-_36237656 | 2.35 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr19_+_20209561 | 2.35 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr14_+_4489377 | 2.34 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr16_+_21121428 | 2.33 |
|
|
|
| chr16_+_14817799 | 2.31 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr6_+_4712785 | 2.30 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr11_-_5868257 | 2.29 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr23_+_2391223 | 2.23 |
|
|
|
| chr18_+_45893199 | 2.23 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr3_+_33168814 | 2.21 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr13_-_24316940 | 2.16 |
ENSDART00000127741
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr24_-_6868340 | 2.13 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr17_+_27417635 | 2.13 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr6_-_14821305 | 2.07 |
|
|
|
| chr7_-_8128948 | 1.95 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr4_-_2615160 | 1.94 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr2_+_42874975 | 1.94 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr11_+_40652239 | 1.91 |
ENSDART00000169817
ENSDART00000162157 ENSDART00000172008 |
pax7a
|
paired box 7a |
| chr13_+_23152038 | 1.91 |
ENSDART00000171676
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr21_+_20678383 | 1.91 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr15_-_31687 | 1.87 |
|
|
|
| chr23_+_10211543 | 1.81 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr12_-_26314881 | 1.77 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr7_+_42124857 | 1.75 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| KN150074v1_-_1031 | 1.75 |
|
|
|
| chr25_+_24520476 | 1.74 |
|
|
|
| chr2_-_36058327 | 1.74 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr16_+_14817718 | 1.74 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr12_+_9664920 | 1.73 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr17_+_52736192 | 1.72 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr25_+_20118286 | 1.71 |
ENSDART00000104297
|
tnnt2d
|
troponin T2d, cardiac |
| chr13_+_30781652 | 1.69 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr17_-_52735250 | 1.66 |
|
|
|
| chr4_-_19027117 | 1.66 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr24_-_4941954 | 1.65 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr16_-_54638972 | 1.64 |
|
|
|
| chr8_-_34077387 | 1.63 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_+_37113743 | 1.62 |
|
|
|
| chr5_+_44407763 | 1.62 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr3_+_30790513 | 1.60 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr5_+_49103778 | 1.58 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr16_+_24032160 | 1.58 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr11_+_7518953 | 1.58 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr9_+_25964943 | 1.56 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_21122024 | 1.56 |
|
|
|
| chr20_-_42805703 | 1.51 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr15_-_33688186 | 1.50 |
|
|
|
| chr21_-_28883441 | 1.48 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr13_+_22350043 | 1.46 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr16_-_19154920 | 1.46 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr3_-_34363698 | 1.46 |
|
|
|
| chr1_+_16983775 | 1.45 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr20_+_19613133 | 1.44 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr18_-_14891913 | 1.42 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr24_+_24308055 | 1.41 |
|
|
|
| chr5_-_18457877 | 1.39 |
ENSDART00000142531
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr16_+_10885785 | 1.37 |
ENSDART00000161969
|
atp1a3b
|
ATPase, Na+/K+ transporting, alpha 3b polypeptide |
| chr13_-_11404389 | 1.37 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr23_+_35819625 | 1.36 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr7_-_33558939 | 1.36 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr1_-_46292847 | 1.35 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr10_+_31358236 | 1.33 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr23_-_6707246 | 1.33 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr15_+_21231406 | 1.30 |
ENSDART00000038499
|
CR626907.1
|
ENSDARG00000077872 |
| chr10_-_26782374 | 1.30 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr5_+_53178362 | 1.29 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr24_-_12794564 | 1.28 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr7_+_26357875 | 1.28 |
ENSDART00000101044
|
hsbp1a
|
heat shock factor binding protein 1a |
| chr1_-_25600988 | 1.28 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr5_+_34857986 | 1.27 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr13_+_30781718 | 1.25 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| KN149932v1_+_27584 | 1.25 |
|
|
|
| chr9_-_20562293 | 1.23 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_-_9387290 | 1.20 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr7_-_39170016 | 1.20 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr6_+_24299180 | 1.18 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr7_+_20251345 | 1.15 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr3_+_23638277 | 1.15 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr16_+_24058139 | 1.14 |
|
|
|
| chr14_+_21999898 | 1.14 |
|
|
|
| chr21_+_22386643 | 1.14 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr21_+_22808694 | 1.14 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr4_-_16344954 | 1.13 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr23_+_43284714 | 1.12 |
|
|
|
| chr12_-_26339002 | 1.11 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr16_+_51319421 | 1.10 |
|
|
|
| chr25_-_13737344 | 1.10 |
|
|
|
| chr4_+_13811932 | 1.09 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr13_-_25067585 | 1.09 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr22_+_16509286 | 1.09 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr23_+_17294623 | 1.09 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr7_+_44269171 | 1.08 |
ENSDART00000149981
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr7_+_45980684 | 1.08 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr13_-_5440751 | 1.08 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr5_-_62747812 | 1.08 |
|
|
|
| chr25_-_3853819 | 1.07 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr12_-_48948928 | 1.07 |
ENSDART00000163734
|
rgrb
|
retinal G protein coupled receptor b |
| chr8_+_18552850 | 1.07 |
ENSDART00000177476
|
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr4_-_998370 | 1.06 |
|
|
|
| chr3_-_50386208 | 1.04 |
ENSDART00000156709
|
BX088529.1
|
ENSDARG00000097178 |
| chr2_+_57903155 | 1.04 |
|
|
|
| chr13_-_5440558 | 1.04 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr21_-_43083936 | 1.04 |
ENSDART00000040169
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr15_+_31036717 | 1.03 |
ENSDART00000129474
|
ogmb
|
oligodendrocyte myelin glycoprotein b |
| chr23_+_16962770 | 1.03 |
ENSDART00000146895
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr5_+_296856 | 1.02 |
|
|
|
| chr6_+_15141723 | 1.02 |
ENSDART00000128090
|
ecrg4b
|
esophageal cancer related gene 4b |
| chr19_-_10324632 | 1.02 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr6_-_10991748 | 1.02 |
ENSDART00000151252
|
myo10l1
|
myosin X-like 1 |
| chr8_-_25055333 | 0.99 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr9_-_23406315 | 0.98 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr4_+_828545 | 0.98 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr21_+_43674754 | 0.98 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr7_+_26770097 | 0.97 |
ENSDART00000173822
|
rps13
|
ribosomal protein S13 |
| chr8_+_22910147 | 0.97 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr9_+_3147601 | 0.97 |
ENSDART00000135619
|
ftr52p
|
finTRIM family, member 52, pseudogene |
| chr16_+_51317933 | 0.96 |
ENSDART00000157736
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr17_+_23892232 | 0.96 |
|
|
|
| chr11_+_13470883 | 0.96 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr23_+_36023748 | 0.95 |
|
|
|
| chr24_-_12794672 | 0.95 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr8_+_15216833 | 0.95 |
ENSDART00000141185
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr17_-_19606453 | 0.94 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr11_-_6051096 | 0.94 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr24_-_6048914 | 0.94 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr23_+_35996491 | 0.92 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr21_+_13063614 | 0.92 |
|
|
|
| chr18_+_23891068 | 0.91 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr7_-_33558771 | 0.89 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr19_-_27284726 | 0.89 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr22_-_24286717 | 0.89 |
ENSDART00000161480
|
uts2d
|
urotensin 2 domain containing |
| chr2_-_844040 | 0.87 |
ENSDART00000028159
|
foxf2a
|
forkhead box F2a |
| chr15_-_35802739 | 0.87 |
ENSDART00000174714
|
CR848683.2
|
ENSDARG00000106192 |
| chr19_+_14197118 | 0.86 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr7_+_24770873 | 0.85 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr13_+_16130123 | 0.85 |
ENSDART00000170855
|
mocos
|
molybdenum cofactor sulfurase |
| chr5_-_63422783 | 0.84 |
ENSDART00000083684
|
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr2_-_44330405 | 0.84 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr3_+_40667131 | 0.84 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr15_-_25592228 | 0.83 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr1_+_38669801 | 0.83 |
ENSDART00000084891
ENSDART00000148701 |
tenm3
|
teneurin transmembrane protein 3 |
| chr3_+_23621561 | 0.83 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr24_+_18804086 | 0.82 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_38669902 | 0.82 |
ENSDART00000084891
ENSDART00000148701 |
tenm3
|
teneurin transmembrane protein 3 |
| chr7_+_53485273 | 0.81 |
ENSDART00000158160
ENSDART00000163261 |
neo1a
|
neogenin 1a |
| chr8_-_38168395 | 0.80 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_27301938 | 0.80 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr5_-_7329952 | 0.80 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr16_-_55234426 | 0.79 |
ENSDART00000078887
|
tmem222a
|
transmembrane protein 222a |
| chr4_+_26507297 | 0.79 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr20_-_7303804 | 0.79 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr19_+_20194594 | 0.78 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| chr1_-_22867130 | 0.78 |
ENSDART00000145942
ENSDART00000077394 |
fam184b
|
family with sequence similarity 184, member B |
| chr24_-_24306469 | 0.77 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr5_-_62747914 | 0.77 |
|
|
|
| chr3_+_23621843 | 0.77 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr19_+_20202737 | 0.76 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr22_-_29886668 | 0.76 |
ENSDART00000125017
|
BX649294.1
|
ENSDARG00000092881 |
| chr15_+_28269289 | 0.76 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr8_-_34077790 | 0.76 |
|
|
|
| chr8_+_5222065 | 0.76 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr3_+_37433008 | 0.76 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr15_+_14048904 | 0.75 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr14_+_5078937 | 0.74 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr24_+_34227202 | 0.74 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr3_+_26015378 | 0.74 |
|
|
|
| chr12_-_28766713 | 0.73 |
ENSDART00000148459
|
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr9_-_30553051 | 0.72 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr5_+_64411412 | 0.72 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr4_-_25496215 | 0.71 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| KN150623v1_+_577 | 0.71 |
|
|
|
| chr21_+_2048601 | 0.71 |
|
|
|
| chr5_-_32687349 | 0.71 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr3_-_62143464 | 0.71 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr18_+_23891152 | 0.70 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr7_-_20201693 | 0.70 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr3_+_15656123 | 0.69 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr4_+_13453879 | 0.69 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr8_-_23744125 | 0.68 |
ENSDART00000141871
|
INAVA
|
innate immunity activator |
| chr24_+_37692365 | 0.68 |
|
|
|
| chr18_+_9228336 | 0.68 |
|
|
|
| chr9_+_25964868 | 0.68 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr23_+_18565721 | 0.68 |
|
|
|
| chr23_-_45646407 | 0.68 |
ENSDART00000149291
|
ENSDARG00000036894
|
ENSDARG00000036894 |
| chr23_-_28286971 | 0.67 |
|
|
|
| chr22_+_20436009 | 0.67 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr1_+_45843326 | 0.67 |
|
|
|
| chr17_+_50622804 | 0.66 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr9_-_12604260 | 0.66 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 1.3 | 3.8 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 1.1 | 4.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.8 | 2.3 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.6 | 1.9 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.6 | 2.2 | GO:0019541 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.5 | 1.6 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.5 | 1.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.5 | 1.9 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.5 | 2.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 1.3 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.4 | 3.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.4 | 2.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.4 | 1.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.5 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.4 | 2.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.4 | 5.3 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.3 | 4.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 1.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.3 | 1.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 2.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 2.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.3 | 1.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.3 | 1.2 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.3 | 1.1 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.3 | 1.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.1 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.2 | 0.7 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.9 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 0.9 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.4 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.2 | 1.0 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.2 | 1.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.3 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 3.6 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.2 | 1.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 1.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.8 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.9 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.1 | 1.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.1 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 1.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.6 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.1 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 2.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 1.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.4 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.9 | GO:0051283 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 2.5 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.2 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.9 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 1.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.5 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 3.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 2.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.7 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0051875 | pigment granule localization(GO:0051875) establishment of pigment granule localization(GO:0051905) |
| 0.0 | 0.2 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 1.8 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.1 | GO:0010887 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 1.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.7 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.4 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.9 | GO:0006814 | sodium ion transport(GO:0006814) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 4.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 3.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 1.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 0.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 3.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 5.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 5.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.7 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.6 | 2.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 3.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.4 | 1.8 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.4 | 2.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.4 | 1.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.4 | 1.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.4 | 2.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.3 | 3.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 1.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.8 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 8.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 1.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 3.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 3.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.1 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 3.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 8.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 4.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 18.6 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 8.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 3.0 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |