Project
DANIO-CODE
Navigation
Downloads
Results for pgr
Z-value: 3.14
Transcription factors associated with pgr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pgr
|
ENSDARG00000035966 | progesterone receptor |
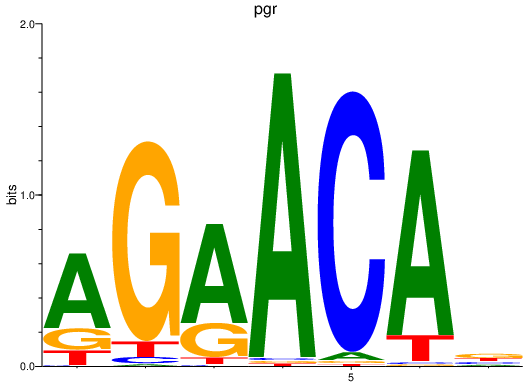
Activity profile of pgr motif
Sorted Z-values of pgr motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pgr
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_14330113 | 9.47 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr20_-_26521700 | 8.57 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr1_-_44922596 | 8.43 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_-_9978112 | 8.11 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr14_-_46648563 | 7.40 |
|
|
|
| chr20_+_33391554 | 7.19 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr13_-_36265504 | 6.76 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr14_-_40454194 | 6.64 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr6_-_47842137 | 6.43 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr5_+_36487425 | 6.25 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr1_-_5048203 | 6.19 |
ENSDART00000150863
ENSDART00000163417 |
nrp2a
|
neuropilin 2a |
| chr3_-_39346621 | 6.03 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr7_+_60992913 | 6.01 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr19_-_19888074 | 5.98 |
|
|
|
| chr1_+_8605984 | 5.97 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr18_+_9213713 | 5.84 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr1_+_38834751 | 5.61 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr4_+_15870605 | 5.59 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr9_+_3458086 | 5.53 |
ENSDART00000160977
ENSDART00000114168 |
CU469503.1
itga6a
|
ENSDARG00000099348 integrin, alpha 6a |
| chr23_-_511984 | 5.49 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr24_-_8589270 | 5.48 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr20_-_29580624 | 5.45 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr4_-_16417703 | 5.42 |
ENSDART00000013085
|
dcn
|
decorin |
| chr10_+_38568464 | 5.23 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr24_-_33869817 | 5.19 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr19_-_47994946 | 5.11 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr16_-_36022327 | 5.08 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr14_+_6117282 | 4.74 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr9_-_33296340 | 4.73 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr18_+_22617072 | 4.72 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr11_-_11925832 | 4.71 |
|
|
|
| chr4_-_14330048 | 4.63 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr5_-_40707316 | 4.59 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr9_-_42894582 | 4.58 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr7_-_32888309 | 4.58 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr18_+_9413588 | 4.56 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr7_+_40604000 | 4.55 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr8_-_24991984 | 4.42 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr14_-_46649324 | 4.33 |
|
|
|
| chr17_+_1610578 | 4.29 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr11_+_21749658 | 4.25 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr1_-_46047321 | 4.24 |
|
|
|
| chr24_-_2278409 | 4.21 |
|
|
|
| chr8_-_14142049 | 4.10 |
ENSDART00000126432
|
rhoaa
|
ras homolog gene family, member Aa |
| chr11_-_4216204 | 4.03 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr5_+_39619826 | 3.99 |
ENSDART00000114078
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr12_-_4497094 | 3.98 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr2_+_26632673 | 3.89 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr4_+_17428131 | 3.86 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr20_+_29731714 | 3.84 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr5_+_10789834 | 3.84 |
|
|
|
| chr20_-_8431338 | 3.82 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr9_+_4410206 | 3.80 |
|
|
|
| chr5_+_22006860 | 3.77 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr12_-_35590747 | 3.76 |
|
|
|
| chr20_-_30474196 | 3.60 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| KN149883v1_-_5164 | 3.60 |
ENSDART00000158506
ENSDART00000172360 |
NDUFA13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr25_+_31769985 | 3.59 |
|
|
|
| chr5_-_43121949 | 3.59 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr3_-_29846530 | 3.57 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_+_9426040 | 3.55 |
ENSDART00000080576
|
lratb
|
lecithin retinol acyltransferase b (phosphatidylcholine-retinol O-acyltransferase) |
| chr3_-_16100715 | 3.54 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_-_67233396 | 3.50 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr9_-_56292487 | 3.50 |
ENSDART00000151720
|
si:ch211-39i22.1
|
si:ch211-39i22.1 |
| chr18_-_6494014 | 3.48 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr25_-_21692269 | 3.48 |
|
|
|
| chr5_-_51109419 | 3.47 |
ENSDART00000163464
|
lhfpl2b
|
lipoma HMGIC fusion partner-like 2b |
| chr6_-_19928053 | 3.46 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr6_+_2849460 | 3.46 |
|
|
|
| chr8_+_23071884 | 3.44 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr14_-_46648801 | 3.42 |
|
|
|
| chr20_-_11111340 | 3.41 |
|
|
|
| chr5_-_58269742 | 3.41 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr5_+_31745031 | 3.39 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr17_+_1058137 | 3.38 |
|
|
|
| chr20_-_8431368 | 3.38 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr9_+_49054601 | 3.34 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr14_-_46649039 | 3.31 |
|
|
|
| chr5_-_62816208 | 3.31 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr2_+_3864843 | 3.31 |
ENSDART00000108964
ENSDART00000128514 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr6_+_24717985 | 3.22 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr14_-_46649231 | 3.16 |
|
|
|
| chr23_+_7445760 | 3.14 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr24_+_36746202 | 3.13 |
ENSDART00000159017
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr6_-_39767452 | 3.12 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr15_-_15532980 | 3.12 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr2_+_47727856 | 3.11 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr22_-_15930756 | 3.10 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr7_+_28862183 | 3.10 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr14_-_46649445 | 3.09 |
|
|
|
| chr18_-_12484760 | 3.09 |
ENSDART00000135574
|
AL929229.1
|
ENSDARG00000091975 |
| chr9_+_17298410 | 3.09 |
ENSDART00000048548
|
scel
|
sciellin |
| chr25_-_12107407 | 3.09 |
ENSDART00000159800
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr1_-_25704525 | 3.09 |
ENSDART00000171614
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr5_-_29858748 | 3.08 |
|
|
|
| chr22_-_951880 | 3.08 |
ENSDART00000105895
ENSDART00000172206 |
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr20_+_6640497 | 3.03 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr12_-_1931281 | 3.02 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr13_+_28364942 | 3.01 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr20_+_23599157 | 2.99 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr13_-_36265476 | 2.96 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr13_-_3801122 | 2.95 |
|
|
|
| chr6_+_40876664 | 2.94 |
ENSDART00000004295
|
zgc:112163
|
zgc:112163 |
| chr18_-_41660821 | 2.94 |
ENSDART00000024087
|
fzd9b
|
frizzled class receptor 9b |
| chr11_-_998023 | 2.89 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr18_+_22120282 | 2.88 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr14_+_23220869 | 2.88 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr21_+_28441951 | 2.88 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr4_+_45204456 | 2.86 |
ENSDART00000158744
|
BX927336.3
|
ENSDARG00000102848 |
| chr14_-_27703812 | 2.82 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr2_+_33399405 | 2.81 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_35135414 | 2.79 |
ENSDART00000140563
|
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr17_-_32913432 | 2.78 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr7_-_44332679 | 2.74 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr23_-_18958008 | 2.74 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr13_+_28365036 | 2.73 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr15_-_12484651 | 2.71 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr15_-_254261 | 2.66 |
|
|
|
| chr25_+_5856023 | 2.63 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr15_-_24934442 | 2.62 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr9_-_47041677 | 2.62 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr8_-_15192275 | 2.61 |
ENSDART00000141763
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr11_-_997918 | 2.57 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr2_+_29992879 | 2.55 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr3_+_27684309 | 2.54 |
|
|
|
| KN150487v1_+_15409 | 2.52 |
ENSDART00000166996
|
CABZ01074304.1
|
ENSDARG00000100224 |
| chr11_+_666006 | 2.51 |
ENSDART00000130839
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr14_-_2131861 | 2.50 |
ENSDART00000162342
|
BX005294.2
|
protocadherin 2 gamma 10 precursor |
| chr5_+_23675413 | 2.49 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr10_-_31838886 | 2.48 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr11_+_27721622 | 2.47 |
ENSDART00000147984
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr5_+_64226125 | 2.45 |
ENSDART00000122863
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr23_+_35996491 | 2.44 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr13_-_31491759 | 2.44 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr19_+_12315183 | 2.42 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr18_+_30391910 | 2.42 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr2_+_309237 | 2.42 |
ENSDART00000156282
|
CU928013.1
|
ENSDARG00000097884 |
| chr20_-_28981879 | 2.41 |
ENSDART00000134564
ENSDART00000132127 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr6_+_1873957 | 2.41 |
|
|
|
| chr24_-_38928988 | 2.39 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr6_-_40101104 | 2.38 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr17_+_25659010 | 2.37 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr5_-_49091552 | 2.37 |
ENSDART00000140464
|
CR751602.1
|
ENSDARG00000094128 |
| chr15_-_25582891 | 2.33 |
|
|
|
| chr19_-_19164098 | 2.32 |
ENSDART00000168004
|
rps18
|
ribosomal protein S18 |
| chr15_-_17933972 | 2.29 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr8_-_46313975 | 2.27 |
ENSDART00000075189
|
mtor
|
mechanistic target of rapamycin (serine/threonine kinase) |
| chr23_-_28286971 | 2.25 |
|
|
|
| chr24_+_14302079 | 2.25 |
ENSDART00000168059
|
prdm14
|
PR domain containing 14 |
| chr15_+_28550096 | 2.23 |
ENSDART00000154320
|
ankrd13b
|
ankyrin repeat domain 13B |
| chr11_-_33656483 | 2.21 |
ENSDART00000078202
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr7_+_24762755 | 2.21 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr16_+_17061069 | 2.20 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr22_-_15576473 | 2.15 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr16_+_38169001 | 2.15 |
ENSDART00000132087
|
pogzb
|
pogo transposable element derived with ZNF domain b |
| chr12_-_36138709 | 2.10 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr16_-_29599005 | 2.09 |
|
|
|
| chr5_+_39066996 | 2.09 |
ENSDART00000085388
|
bmp3
|
bone morphogenetic protein 3 |
| chr5_+_39067074 | 2.09 |
ENSDART00000085388
|
bmp3
|
bone morphogenetic protein 3 |
| chr2_-_37914464 | 2.07 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr8_-_39950775 | 2.00 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr19_+_14669633 | 2.00 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr17_+_16557298 | 2.00 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr19_+_34145030 | 2.00 |
ENSDART00000151521
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_+_6070537 | 1.95 |
ENSDART00000123797
|
pcp4b
|
Purkinje cell protein 4b |
| chr9_-_47042015 | 1.95 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr5_-_24365811 | 1.94 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr14_+_24543732 | 1.93 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr9_-_37940101 | 1.91 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr10_+_213828 | 1.90 |
ENSDART00000138812
|
mpzl1l
|
myelin protein zero-like 1 like |
| chr18_+_27205666 | 1.88 |
|
|
|
| chr2_-_32468715 | 1.87 |
|
|
|
| chr1_-_44922521 | 1.87 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_-_13890202 | 1.86 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr18_-_40518772 | 1.82 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr14_-_2274883 | 1.82 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr16_-_31670211 | 1.79 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr16_-_16796313 | 1.79 |
|
|
|
| chr4_-_16344954 | 1.77 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr3_+_32566693 | 1.77 |
ENSDART00000075244
|
fus
|
FUS RNA binding protein |
| chr19_+_32679320 | 1.77 |
|
|
|
| chr14_-_25688444 | 1.76 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr24_+_11194381 | 1.75 |
ENSDART00000143171
|
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr10_-_30016761 | 1.73 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr1_-_8968543 | 1.72 |
ENSDART00000126877
ENSDART00000123773 ENSDART00000126996 |
ugt5b1
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B1 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr18_-_47186938 | 1.72 |
|
|
|
| chr21_-_10354049 | 1.70 |
ENSDART00000159958
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr10_+_1954904 | 1.69 |
ENSDART00000177433
ENSDART00000178960 |
CABZ01070746.1
|
ENSDARG00000107761 |
| chr11_-_18068313 | 1.68 |
|
|
|
| chr16_+_5542562 | 1.67 |
|
|
|
| chr19_+_32679258 | 1.67 |
|
|
|
| chr9_-_1978279 | 1.67 |
|
|
|
| chr10_-_37098396 | 1.65 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr9_-_22461196 | 1.65 |
ENSDART00000135190
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr6_-_39313219 | 1.63 |
|
|
|
| chr3_-_29846789 | 1.62 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_-_29849858 | 1.62 |
ENSDART00000052108
ENSDART00000171908 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr13_-_36409205 | 1.61 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr10_+_9134634 | 1.61 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr6_+_54931721 | 1.60 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0061193 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 1.9 | 5.8 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 1.9 | 7.7 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 1.9 | 5.6 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.8 | 7.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.4 | 5.7 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 1.3 | 4.0 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 1.2 | 4.7 | GO:0070296 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 1.1 | 5.4 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 1.0 | 6.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.9 | 4.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.9 | 3.7 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.9 | 4.6 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.9 | 4.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.9 | 10.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 2.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.8 | 4.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.8 | 2.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.8 | 3.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 2.8 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.7 | 3.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.6 | 4.7 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.6 | 11.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 3.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.6 | 2.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.6 | 3.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 2.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.5 | 4.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 3.1 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 3.6 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.4 | 6.5 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.4 | 3.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.4 | 2.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.4 | 1.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 2.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.4 | 1.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 3.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 2.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 2.4 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.3 | 3.1 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.3 | 6.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.3 | 1.3 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.3 | 6.1 | GO:0014855 | striated muscle cell proliferation(GO:0014855) cardiac muscle cell proliferation(GO:0060038) |
| 0.3 | 5.4 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.3 | 6.5 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.3 | 3.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.3 | 1.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 5.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 1.8 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 1.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 2.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 0.5 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 3.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.2 | 4.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.2 | 2.4 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.2 | 1.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.2 | 1.3 | GO:0097028 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.2 | 1.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 6.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 4.0 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.1 | 3.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 3.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.1 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 3.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 9.7 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.1 | 2.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 6.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.3 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 11.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 2.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.0 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.1 | 0.9 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 2.5 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 1.2 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.0 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 2.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 3.6 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.6 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.6 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.2 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 2.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.4 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 3.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.0 | 1.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.0 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.8 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 2.8 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 3.8 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 4.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 3.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.5 | 3.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 2.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 3.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 3.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 5.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 3.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 6.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 9.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 3.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 3.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 15.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 3.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 15.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 3.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 32.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.5 | GO:0030665 | clathrin vesicle coat(GO:0030125) clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 8.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.3 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 4.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 9.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 14.4 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.0 | 14.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.8 | 3.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.8 | 2.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.8 | 4.7 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.8 | 5.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.8 | 3.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.7 | 10.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.7 | 4.4 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.7 | 6.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 2.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) organic acid:sodium symporter activity(GO:0005343) |
| 0.5 | 10.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.5 | 4.6 | GO:0031995 | fibronectin binding(GO:0001968) insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 4.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 1.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 3.1 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 3.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 1.2 | GO:0043121 | neurotrophin binding(GO:0043121) nerve growth factor binding(GO:0048406) |
| 0.4 | 3.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 3.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 3.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 4.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.3 | 3.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 4.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.8 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 5.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 1.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 4.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 2.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 1.1 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 3.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.2 | 2.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 10.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.1 | GO:0016493 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 3.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 5.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 6.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.2 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.1 | 2.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 14.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 5.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 3.5 | GO:0031406 | carboxylic acid binding(GO:0031406) |
| 0.0 | 20.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 3.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.0 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 6.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 23.9 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.7 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.6 | 10.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.5 | 8.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 8.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 6.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 2.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 2.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 9.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 7.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 3.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 6.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 1.8 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.3 | 8.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 2.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 1.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 4.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |