Project
DANIO-CODE
Navigation
Downloads
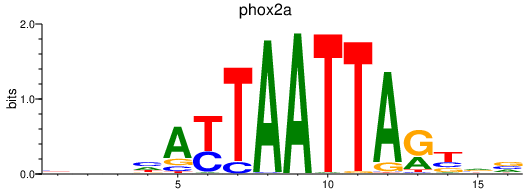
Results for phox2a
Z-value: 0.42
Transcription factors associated with phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2a
|
ENSDARG00000007406 | paired like homeobox 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2a | dr10_dc_chr15_+_46976241_46976246 | -0.79 | 2.7e-04 | Click! |
Activity profile of phox2a motif
Sorted Z-values of phox2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34058331 | 1.27 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr12_-_33256671 | 1.19 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_+_37303599 | 1.15 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_-_33256754 | 1.08 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr24_+_19270877 | 0.98 |
|
|
|
| chr12_-_33257026 | 0.92 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_-_33256599 | 0.89 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33257120 | 0.80 |
|
|
|
| chr14_-_6901209 | 0.70 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr21_-_32027717 | 0.69 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr12_-_33256934 | 0.67 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_26941232 | 0.65 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_-_26941084 | 0.62 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_-_26940965 | 0.48 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr20_-_29961498 | 0.48 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_-_29961589 | 0.41 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr14_-_6901783 | 0.39 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr21_-_13026036 | 0.38 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr17_+_8642686 | 0.30 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_+_29601073 | 0.28 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr21_+_43333462 | 0.27 |
ENSDART00000109620
|
sept8a
|
septin 8a |
| chr21_-_13026077 | 0.27 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_+_37303707 | 0.26 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr17_+_8642373 | 0.26 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr20_+_35176069 | 0.22 |
|
|
|
| chr24_+_16402587 | 0.18 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr6_+_55250892 | 0.17 |
|
|
|
| chr15_-_43402935 | 0.16 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr4_-_1952230 | 0.14 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr14_+_29601252 | 0.13 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr3_-_26657213 | 0.12 |
|
|
|
| chr16_+_51597409 | 0.07 |
|
|
|
| chr7_+_28814408 | 0.04 |
ENSDART00000173960
|
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr18_+_1353857 | 0.00 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 1.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |