Project
DANIO-CODE
Navigation
Downloads
Results for phox2bb
Z-value: 0.31
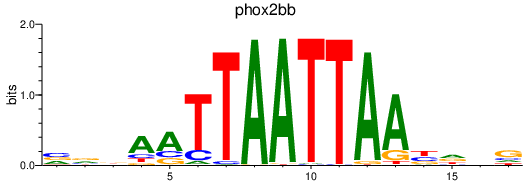
Transcription factors associated with phox2bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2bb
|
ENSDARG00000091029 | paired like homeobox 2Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2bb | dr10_dc_chr14_-_16762869_16762893 | 0.65 | 6.6e-03 | Click! |
Activity profile of phox2bb motif
Sorted Z-values of phox2bb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2bb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_9107294 | 0.82 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr21_+_25199691 | 0.80 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr17_+_15425559 | 0.72 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr8_-_33372638 | 0.70 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr16_+_46145286 | 0.60 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr6_-_43094573 | 0.58 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr15_-_14616083 | 0.57 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr9_-_37940101 | 0.50 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr1_+_13779755 | 0.43 |
|
|
|
| chr1_-_30801520 | 0.43 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr12_-_7902815 | 0.38 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr6_-_43094926 | 0.33 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr14_+_2969679 | 0.32 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr7_+_25052687 | 0.26 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr11_-_40414239 | 0.24 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr2_+_38390887 | 0.21 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr9_-_43736549 | 0.19 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr1_-_9256864 | 0.18 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr8_-_12253026 | 0.17 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr2_-_17721575 | 0.17 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr4_-_1952201 | 0.16 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr3_+_3994248 | 0.16 |
|
|
|
| chr10_-_27087472 | 0.15 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr20_+_109649 | 0.14 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr6_+_11014565 | 0.12 |
ENSDART00000132677
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr2_+_50873893 | 0.09 |
|
|
|
| chr7_+_21006803 | 0.09 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr22_+_12306260 | 0.07 |
|
|
|
| chr11_-_40414131 | 0.07 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr6_+_40924749 | 0.07 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_26657213 | 0.06 |
|
|
|
| chr18_+_1353857 | 0.05 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr17_-_16414629 | 0.05 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr13_+_33138144 | 0.04 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr1_+_9634016 | 0.03 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr2_+_1881022 | 0.03 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr14_-_33604914 | 0.03 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr1_+_9633924 | 0.02 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr9_-_32366320 | 0.02 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr14_-_1187074 | 0.01 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr17_-_21398340 | 0.01 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr2_+_50873843 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |