Project
DANIO-CODE
Navigation
Downloads
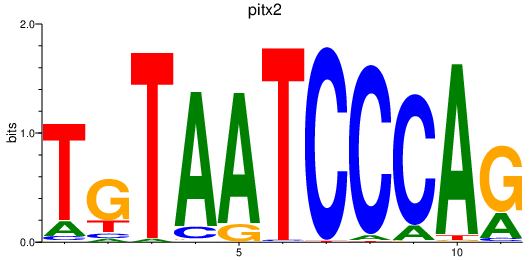
Results for pitx2
Z-value: 1.21
Transcription factors associated with pitx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx2
|
ENSDARG00000036194 | paired-like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx2 | dr10_dc_chr14_+_35885903_35885994 | 0.65 | 6.0e-03 | Click! |
Activity profile of pitx2 motif
Sorted Z-values of pitx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_46356213 | 2.81 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr17_+_1080921 | 2.78 |
|
|
|
| chr15_+_35184996 | 2.35 |
ENSDART00000086954
ENSDART00000161594 |
sesn3
|
sestrin 3 |
| chr12_+_30591157 | 2.10 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr6_+_29226868 | 1.97 |
ENSDART00000006386
|
atp1b1a
|
ATPase, Na+/K+ transporting, beta 1a polypeptide |
| chr15_-_38027363 | 1.86 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr14_+_6117282 | 1.86 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr14_-_40454194 | 1.77 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_-_25649570 | 1.75 |
ENSDART00000154733
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr16_+_46728964 | 1.65 |
ENSDART00000163571
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr8_-_31098193 | 1.59 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr14_-_5239605 | 1.59 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr3_-_10342940 | 1.53 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr12_+_9504247 | 1.49 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr8_+_17148864 | 1.39 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr23_+_23305483 | 1.37 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr11_+_5929544 | 1.30 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr19_+_17832515 | 1.29 |
ENSDART00000149045
|
klf2b
|
Kruppel-like factor 2b |
| chr7_-_54407461 | 1.21 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr11_+_6446302 | 1.11 |
ENSDART00000140707
ENSDART00000036939 |
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr5_+_45822196 | 1.10 |
|
|
|
| chr7_+_23843682 | 1.10 |
ENSDART00000173899
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr15_+_46114986 | 1.09 |
ENSDART00000156366
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr17_-_25865861 | 1.09 |
|
|
|
| chr16_-_45943282 | 1.05 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr19_-_28699796 | 1.04 |
ENSDART00000079104
|
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr23_+_18565721 | 1.03 |
|
|
|
| chr15_-_643531 | 0.98 |
ENSDART00000102257
|
alox5b.3
|
arachidonate 5-lipoxygenase b, tandem duplicate 3 |
| chr16_-_17350056 | 0.97 |
ENSDART00000079497
|
emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr8_-_22486746 | 0.91 |
ENSDART00000101616
|
si:ch211-261n11.5
|
si:ch211-261n11.5 |
| chr13_+_43266551 | 0.88 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr7_+_38455040 | 0.87 |
|
|
|
| chr7_+_73598802 | 0.86 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr2_+_25468904 | 0.86 |
ENSDART00000056801
|
msl2a
|
male-specific lethal 2 homolog a (Drosophila) |
| chr25_+_21732255 | 0.83 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_+_63068121 | 0.83 |
ENSDART00000135097
|
si:ch73-274k23.1
|
si:ch73-274k23.1 |
| chr19_-_15951003 | 0.82 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| KN149859v1_+_16751 | 0.82 |
ENSDART00000167610
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| chr6_+_29226824 | 0.80 |
ENSDART00000006386
|
atp1b1a
|
ATPase, Na+/K+ transporting, beta 1a polypeptide |
| chr18_-_44323435 | 0.79 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr5_+_58017820 | 0.78 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
| chr5_+_11340748 | 0.77 |
ENSDART00000159248
|
tesca
|
tescalcin a |
| chr7_+_37105966 | 0.75 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr20_-_32431934 | 0.73 |
|
|
|
| chr8_+_21974863 | 0.69 |
|
|
|
| chr21_+_13030816 | 0.68 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr15_-_20088439 | 0.67 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr3_-_24550538 | 0.65 |
ENSDART00000161612
|
BX569789.1
|
ENSDARG00000099678 |
| chr10_-_43924675 | 0.65 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr10_+_17470158 | 0.61 |
|
|
|
| chr25_-_17271237 | 0.60 |
ENSDART00000110692
|
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr23_+_8261065 | 0.60 |
ENSDART00000044733
|
NPBWR2 (1 of many)
|
neuropeptides B and W receptor 2 |
| chr19_-_32214978 | 0.60 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr8_-_23591082 | 0.56 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr16_+_26989962 | 0.56 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr11_-_4216204 | 0.56 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr3_+_50398735 | 0.55 |
|
|
|
| chr24_-_16995194 | 0.54 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr21_+_13031158 | 0.54 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr25_+_34515221 | 0.54 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr22_+_1296143 | 0.54 |
ENSDART00000170421
|
si:ch73-138e16.6
|
si:ch73-138e16.6 |
| chr15_+_35185174 | 0.53 |
ENSDART00000086954
ENSDART00000161594 |
sesn3
|
sestrin 3 |
| chr6_+_24211657 | 0.53 |
|
|
|
| chr4_-_29003925 | 0.51 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr23_+_22649686 | 0.51 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr16_-_25748690 | 0.51 |
ENSDART00000031304
|
derl1
|
derlin 1 |
| chr23_+_9285446 | 0.49 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr8_+_17148832 | 0.49 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr23_-_45927723 | 0.48 |
|
|
|
| chr10_+_17470092 | 0.47 |
|
|
|
| chr11_-_22211478 | 0.47 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr23_-_17078246 | 0.46 |
ENSDART00000126841
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr17_+_25503946 | 0.46 |
|
|
|
| chr25_-_35817806 | 0.45 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr2_+_3125365 | 0.45 |
ENSDART00000164308
ENSDART00000167649 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr4_-_17226968 | 0.44 |
ENSDART00000177956
ENSDART00000177406 |
BX547928.1
|
ENSDARG00000106175 |
| chr19_+_44374082 | 0.44 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr1_-_29958288 | 0.42 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr16_+_25749018 | 0.39 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr18_-_46355957 | 0.39 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr6_-_6160613 | 0.38 |
|
|
|
| chr13_-_11873376 | 0.38 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr21_-_40389601 | 0.38 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr24_-_21114505 | 0.36 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr9_-_23936295 | 0.35 |
ENSDART00000165907
|
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr25_-_34634503 | 0.35 |
ENSDART00000157370
|
CR762436.3
|
Histone H3.2 |
| chr23_+_23305376 | 0.34 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr18_+_618393 | 0.34 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr23_-_18454945 | 0.33 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr16_-_6909034 | 0.33 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr3_+_42217445 | 0.33 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr21_+_13031206 | 0.32 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr16_-_51087813 | 0.32 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr15_-_38027496 | 0.32 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr10_+_17470024 | 0.32 |
|
|
|
| chr24_-_18341006 | 0.30 |
ENSDART00000172533
|
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr1_-_11707288 | 0.30 |
ENSDART00000134708
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr2_+_14805305 | 0.30 |
|
|
|
| chr3_+_25888520 | 0.29 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr21_+_43715322 | 0.29 |
ENSDART00000115799
|
SNORD83
|
Small nucleolar RNA SNORD83 |
| chr7_+_53143741 | 0.28 |
ENSDART00000164643
|
ENSDARG00000104530
|
ENSDARG00000104530 |
| chr7_+_21592861 | 0.28 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr24_+_17044956 | 0.27 |
|
|
|
| chr6_+_52242112 | 0.27 |
ENSDART00000041549
|
zc3h10
|
zinc finger CCCH-type containing 10 |
| chr3_+_30117895 | 0.27 |
ENSDART00000122203
|
Metazoa_SRP
|
Metazoan signal recognition particle RNA |
| chr16_+_12922721 | 0.27 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr19_-_28699617 | 0.27 |
ENSDART00000151756
|
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr5_+_3375612 | 0.27 |
|
|
|
| chr1_+_39965440 | 0.27 |
ENSDART00000127907
|
wu:fc34e06
|
wu:fc34e06 |
| chr2_-_6380457 | 0.26 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr2_+_58654570 | 0.26 |
ENSDART00000164102
ENSDART00000158777 |
cirbpa
|
cold inducible RNA binding protein a |
| chr22_+_5657904 | 0.25 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr7_-_25991743 | 0.25 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr6_-_12787356 | 0.25 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr8_-_38308535 | 0.25 |
|
|
|
| chr19_-_40599163 | 0.24 |
ENSDART00000150972
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr9_+_32496059 | 0.24 |
ENSDART00000078513
|
mob4
|
MOB family member 4, phocein |
| chr15_-_8343858 | 0.24 |
ENSDART00000137552
|
si:ch211-113p18.3
|
si:ch211-113p18.3 |
| chr19_+_7839888 | 0.24 |
|
|
|
| chr1_+_35058654 | 0.23 |
ENSDART00000131284
|
hhip
|
hedgehog interacting protein |
| chr18_-_40518772 | 0.23 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr23_-_31474216 | 0.22 |
ENSDART00000132736
|
lca5
|
Leber congenital amaurosis 5 |
| chr20_+_32599031 | 0.22 |
ENSDART00000145481
|
sec63
|
SEC63 homolog, protein translocation regulator |
| chr15_+_20428823 | 0.22 |
ENSDART00000062676
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr18_-_12302111 | 0.22 |
ENSDART00000062216
|
nup205
|
nucleoporin 205 |
| chr1_+_6862901 | 0.22 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr10_-_10058065 | 0.22 |
ENSDART00000129151
ENSDART00000137392 |
strbp
|
spermatid perinuclear RNA binding protein |
| chr23_+_22649652 | 0.22 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr17_-_5426195 | 0.21 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr6_+_27072030 | 0.21 |
ENSDART00000149429
|
boka
|
BCL2-related ovarian killer a |
| chr17_+_1058137 | 0.21 |
|
|
|
| chr21_+_11274670 | 0.21 |
ENSDART00000091835
|
gtf3c5
|
general transcription factor IIIC, polypeptide 5 |
| chr7_+_5846555 | 0.20 |
ENSDART00000083397
|
hist1h4l
|
histone 1, H4, like |
| chr3_-_5734028 | 0.20 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr13_+_36459625 | 0.20 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr10_+_17414243 | 0.19 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr18_-_5822255 | 0.19 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr11_-_29316162 | 0.19 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr16_-_25748643 | 0.19 |
ENSDART00000132136
|
derl1
|
derlin 1 |
| chr5_+_23555868 | 0.19 |
ENSDART00000124379
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr8_+_10302071 | 0.18 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr13_+_36459706 | 0.18 |
ENSDART00000138940
|
gmfb
|
glia maturation factor, beta |
| chr9_+_308260 | 0.18 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr11_+_15988149 | 0.18 |
ENSDART00000168083
|
Metazoa_SRP
|
Metazoan signal recognition particle RNA |
| chr2_+_49787853 | 0.18 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr11_-_35788900 | 0.18 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr3_-_33812379 | 0.18 |
|
|
|
| chr14_+_28180174 | 0.17 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr1_+_55077656 | 0.17 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr24_+_12845250 | 0.17 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr13_+_36459562 | 0.16 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr8_-_46963909 | 0.16 |
|
|
|
| chr19_+_35155538 | 0.16 |
ENSDART00000146777
ENSDART00000103276 |
fam206a
|
family with sequence similarity 206, member A |
| chr9_+_26218474 | 0.16 |
ENSDART00000177394
|
CR391949.1
|
ENSDARG00000107108 |
| chr7_+_45748322 | 0.16 |
ENSDART00000169822
|
ccne1
|
cyclin E1 |
| chr2_-_1753058 | 0.16 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr19_+_46631965 | 0.16 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr5_+_64159481 | 0.16 |
ENSDART00000073953
|
lrrc8ab
|
leucine rich repeat containing 8 family, member Ab |
| chr8_-_556380 | 0.16 |
ENSDART00000050352
|
si:ch211-87m7.2
|
si:ch211-87m7.2 |
| chr19_-_5429428 | 0.15 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr8_-_22517872 | 0.15 |
ENSDART00000132680
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr12_-_17013870 | 0.15 |
|
|
|
| chr20_+_32598608 | 0.15 |
ENSDART00000021035
ENSDART00000152944 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr9_-_32942783 | 0.15 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr19_-_22761674 | 0.15 |
|
|
|
| chr23_+_43362722 | 0.15 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr20_-_35343057 | 0.15 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr11_+_5929498 | 0.15 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr5_-_70991958 | 0.14 |
ENSDART00000011955
ENSDART00000160380 |
gpsm1b
|
G protein signaling modulator 1b |
| chr6_+_42340776 | 0.14 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr20_-_50249356 | 0.14 |
ENSDART00000123634
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr11_+_12459272 | 0.14 |
ENSDART00000172127
|
Metazoa_SRP
|
Metazoan signal recognition particle RNA |
| chr19_-_20508993 | 0.14 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr18_-_17527321 | 0.14 |
|
|
|
| chr23_+_17912784 | 0.14 |
ENSDART00000104647
|
prim1
|
primase, DNA, polypeptide 1 |
| chr14_-_33596430 | 0.14 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr3_-_10343029 | 0.13 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr14_-_15393962 | 0.13 |
ENSDART00000161123
ENSDART00000167146 |
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr6_-_6091628 | 0.13 |
ENSDART00000132350
|
rtn4a
|
reticulon 4a |
| chr3_-_24977167 | 0.13 |
ENSDART00000055432
|
rbx1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr12_-_30385893 | 0.12 |
ENSDART00000153108
|
nhlrc2
|
NHL repeat containing 2 |
| chr5_-_22090338 | 0.12 |
|
|
|
| chr13_+_49154065 | 0.12 |
|
|
|
| chr6_+_42340724 | 0.12 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr20_-_27193158 | 0.12 |
ENSDART00000062096
|
si:dkeyp-55f12.3
|
si:dkeyp-55f12.3 |
| chr23_+_44543892 | 0.12 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr2_+_37312637 | 0.11 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr7_+_20008083 | 0.11 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr16_+_38251653 | 0.11 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr3_+_28808901 | 0.11 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr3_-_33811737 | 0.11 |
ENSDART00000026090
ENSDART00000047660 ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr23_-_30114528 | 0.10 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr8_+_16954190 | 0.10 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr15_+_37644266 | 0.10 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr25_+_14302256 | 0.10 |
ENSDART00000130548
|
ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr12_+_30538113 | 0.10 |
ENSDART00000179355
|
thbs2b
|
thrombospondin 2b |
| chr16_+_46728897 | 0.10 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr14_+_596346 | 0.10 |
ENSDART00000178302
|
CABZ01076223.1
|
ENSDARG00000106587 |
| chr23_+_10461899 | 0.09 |
ENSDART00000046577
|
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr13_-_37001997 | 0.09 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.7 | 2.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.4 | 1.3 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.4 | 1.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 1.1 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.4 | 2.9 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 1.0 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.2 | GO:0031571 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 1.9 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 0.9 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 2.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.5 | GO:0071846 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.2 | 1.6 | GO:0008406 | gonad development(GO:0008406) negative regulation of endodermal cell fate specification(GO:0042664) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 0.5 | GO:1902024 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 1.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.8 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 2.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) interferon-gamma-mediated signaling pathway(GO:0060333) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 | 0.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 1.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 2.8 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 2.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.2 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 1.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0031207 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 2.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 1.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 1.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 2.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 1.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 3.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |