Project
DANIO-CODE
Navigation
Downloads
Results for pitx3
Z-value: 0.22
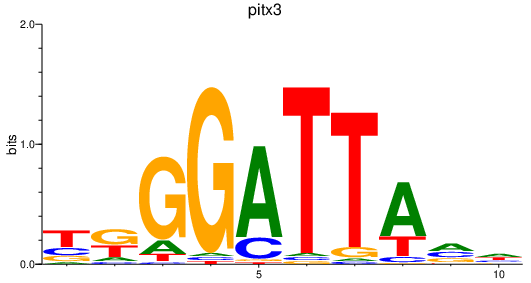
Transcription factors associated with pitx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx3
|
ENSDARG00000070069 | paired-like homeodomain 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx3 | dr10_dc_chr13_-_7241422_7241494 | -0.04 | 8.9e-01 | Click! |
Activity profile of pitx3 motif
Sorted Z-values of pitx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_26013873 | 0.37 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr21_+_27346176 | 0.15 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr21_-_11539798 | 0.14 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr6_-_13910679 | 0.12 |
ENSDART00000065361
|
etv5b
|
ets variant 5b |
| chr7_+_45748011 | 0.12 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr4_-_17226968 | 0.11 |
ENSDART00000177956
ENSDART00000177406 |
BX547928.1
|
ENSDARG00000106175 |
| chr7_+_53143741 | 0.11 |
ENSDART00000164643
|
ENSDARG00000104530
|
ENSDARG00000104530 |
| chr11_-_29316162 | 0.11 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr20_+_29685331 | 0.08 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr23_+_36832325 | 0.08 |
|
|
|
| chr20_+_29685096 | 0.07 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr7_-_66641971 | 0.07 |
|
|
|
| chr18_+_9228336 | 0.07 |
|
|
|
| chr23_+_22649686 | 0.06 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr5_+_45822196 | 0.06 |
|
|
|
| chr18_+_7680416 | 0.06 |
ENSDART00000132369
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr12_-_7902815 | 0.06 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr14_+_6117282 | 0.05 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr7_-_66641207 | 0.05 |
|
|
|
| chr7_+_45748086 | 0.05 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr19_-_39048324 | 0.05 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr5_-_16479187 | 0.04 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr25_-_20932941 | 0.04 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr3_-_26013660 | 0.04 |
|
|
|
| chr25_+_18467034 | 0.04 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr2_-_32521879 | 0.04 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr2_-_32755182 | 0.04 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr17_+_777154 | 0.03 |
ENSDART00000161446
|
si:ch211-193k8.2
|
si:ch211-193k8.2 |
| chr25_+_21735567 | 0.03 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr8_+_23083842 | 0.03 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr9_+_30822402 | 0.03 |
|
|
|
| chr4_+_11465367 | 0.03 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr18_-_5822255 | 0.03 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr21_+_1117194 | 0.03 |
ENSDART00000178294
|
CABZ01086137.1
|
ENSDARG00000107408 |
| chr8_+_23084130 | 0.02 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr3_-_20989024 | 0.02 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr23_+_22649652 | 0.02 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr16_-_51434927 | 0.02 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr8_+_23334921 | 0.02 |
ENSDART00000085361
ENSDART00000046460 ENSDART00000145062 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr6_+_28232386 | 0.02 |
ENSDART00000163057
|
FP101882.1
|
ENSDARG00000098121 |
| chr25_+_18467835 | 0.02 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr16_-_25748690 | 0.02 |
ENSDART00000031304
|
derl1
|
derlin 1 |
| chr6_-_12786937 | 0.02 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr7_-_29262814 | 0.02 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr12_-_19056530 | 0.01 |
|
|
|
| chr19_-_39048402 | 0.01 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr17_+_16038358 | 0.01 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_+_60234640 | 0.01 |
|
|
|
| chr13_-_22653869 | 0.01 |
|
|
|
| chr7_+_53143890 | 0.01 |
ENSDART00000164643
|
ENSDARG00000104530
|
ENSDARG00000104530 |
| chr1_-_40211790 | 0.01 |
ENSDART00000113087
|
hmx1
|
H6 family homeobox 1 |
| chr7_+_45747622 | 0.01 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr6_-_54118302 | 0.01 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr2_-_48317047 | 0.01 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr24_+_17044956 | 0.01 |
|
|
|
| chr5_-_40473026 | 0.01 |
ENSDART00000174936
|
pdzd2
|
PDZ domain containing 2 |
| chr3_-_49104540 | 0.01 |
ENSDART00000138503
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr25_+_18467217 | 0.01 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr19_+_43928043 | 0.01 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr20_+_29685379 | 0.01 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr12_+_27122352 | 0.00 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr17_+_777107 | 0.00 |
ENSDART00000161446
|
si:ch211-193k8.2
|
si:ch211-193k8.2 |
| chr16_+_29651305 | 0.00 |
ENSDART00000124232
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr21_+_5597536 | 0.00 |
ENSDART00000161235
ENSDART00000170456 |
shroom3
|
shroom family member 3 |
| chr7_-_17527827 | 0.00 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045933 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 0.0 | 0.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |