Project
DANIO-CODE
Navigation
Downloads
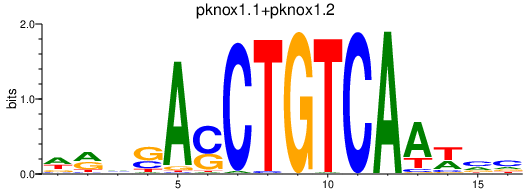
Results for pknox1.1+pknox1.2_meis2b
Z-value: 2.04
Transcription factors associated with pknox1.1+pknox1.2_meis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
|
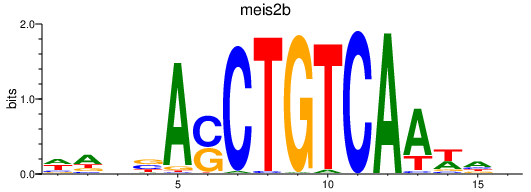
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis2b | dr10_dc_chr20_-_10131480_10131649 | -0.83 | 7.6e-05 | Click! |
| pknox1.2 | dr10_dc_chr1_-_46292847_46292912 | -0.78 | 3.4e-04 | Click! |
| pknox1.1 | dr10_dc_chr9_+_19719018_19719087 | -0.08 | 7.6e-01 | Click! |
Activity profile of pknox1.1+pknox1.2_meis2b motif
Sorted Z-values of pknox1.1+pknox1.2_meis2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pknox1.1+pknox1.2_meis2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_28531087 | 9.31 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr14_-_41101660 | 7.32 |
ENSDART00000003170
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr9_+_4382041 | 6.96 |
|
|
|
| chr8_-_32376710 | 6.90 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr25_+_22222388 | 6.53 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr5_-_12086676 | 5.84 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr4_-_17289857 | 5.66 |
ENSDART00000178686
|
lrmp
|
lymphoid-restricted membrane protein |
| chr13_+_22973764 | 5.25 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr24_+_40770039 | 5.23 |
|
|
|
| chr17_-_36913213 | 5.16 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr19_+_42661987 | 4.10 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr22_-_21021942 | 4.09 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr8_+_25940758 | 4.06 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr17_-_7635061 | 3.91 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr22_-_37899243 | 3.90 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr20_-_28531019 | 3.58 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr14_-_20940726 | 3.54 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr8_+_25940883 | 3.52 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr15_+_545678 | 3.50 |
ENSDART00000102275
ENSDART00000102274 |
ftr86
|
finTRIM family, member 86 |
| chr5_-_12086593 | 3.28 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr15_+_23592828 | 3.14 |
ENSDART00000152422
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr17_+_5680342 | 3.14 |
ENSDART00000156089
|
CU929322.1
|
ENSDARG00000096839 |
| chr5_-_39205454 | 3.07 |
ENSDART00000133231
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr6_+_422911 | 2.95 |
|
|
|
| chr18_+_39506453 | 2.94 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr18_-_49291686 | 2.91 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr24_+_23571714 | 2.91 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr20_+_15072029 | 2.90 |
ENSDART00000063874
|
vamp4
|
vesicle-associated membrane protein 4 |
| chr11_+_29299382 | 2.88 |
|
|
|
| chr7_-_20556760 | 2.83 |
ENSDART00000143509
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr10_+_36752015 | 2.82 |
ENSDART00000171392
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr20_-_46563338 | 2.80 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr25_+_14773723 | 2.79 |
ENSDART00000035714
|
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr15_-_28654137 | 2.75 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr5_-_36503296 | 2.70 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr16_-_32708662 | 2.67 |
ENSDART00000133351
|
coq3
|
coenzyme Q3 methyltransferase |
| chr1_+_57267909 | 2.67 |
ENSDART00000152640
|
BX469930.2
|
ENSDARG00000096615 |
| chr5_-_40524177 | 2.65 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr20_-_35601497 | 2.61 |
|
|
|
| chr12_-_33558248 | 2.59 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr25_+_22222336 | 2.57 |
ENSDART00000154065
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr17_-_36913302 | 2.54 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr10_+_45108698 | 2.45 |
|
|
|
| chr14_-_38549466 | 2.44 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr3_+_16880947 | 2.44 |
ENSDART00000080854
|
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr11_+_8142982 | 2.41 |
ENSDART00000166379
|
samd13
|
sterile alpha motif domain containing 13 |
| chr25_+_14773447 | 2.39 |
ENSDART00000171835
|
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr15_+_545631 | 2.35 |
ENSDART00000102275
ENSDART00000102274 |
ftr86
|
finTRIM family, member 86 |
| chr12_+_18794467 | 2.34 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr20_+_34021257 | 2.33 |
ENSDART00000146292
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr13_-_24265471 | 2.31 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr6_+_49054117 | 2.30 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr4_-_20092577 | 2.29 |
ENSDART00000164410
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr16_-_32708605 | 2.28 |
ENSDART00000133351
|
coq3
|
coenzyme Q3 methyltransferase |
| chr16_-_11981275 | 2.27 |
|
|
|
| chr10_+_37556367 | 2.25 |
ENSDART00000135642
|
msi2a
|
musashi RNA-binding protein 2a |
| chr7_-_25991514 | 2.16 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr11_-_24300905 | 2.14 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr20_-_20370397 | 2.13 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr8_-_41511380 | 2.11 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr24_-_20812363 | 2.06 |
ENSDART00000142080
|
kpna1
|
karyopherin alpha 1 (importin alpha 5) |
| chr2_+_50680126 | 2.01 |
ENSDART00000122716
|
ENSDARG00000090398
|
ENSDARG00000090398 |
| chr3_-_22240424 | 2.01 |
|
|
|
| chr3_-_23466232 | 2.00 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr11_-_15886860 | 2.00 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr4_+_27909339 | 1.98 |
ENSDART00000100453
|
cerk
|
ceramide kinase |
| chr25_+_2252667 | 1.97 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr21_-_23009910 | 1.97 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr20_+_34021116 | 1.96 |
ENSDART00000146292
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr2_-_45657620 | 1.94 |
ENSDART00000056357
|
gpsm2
|
G protein signaling modulator 2 |
| chr7_-_56465369 | 1.93 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr22_-_21021984 | 1.93 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr6_-_39162916 | 1.92 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr5_-_31889551 | 1.92 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr11_+_29295032 | 1.91 |
ENSDART00000112721
|
BX548000.1
|
ENSDARG00000079546 |
| chr12_+_23543100 | 1.90 |
ENSDART00000111334
|
mtpap
|
mitochondrial poly(A) polymerase |
| chr5_-_22981395 | 1.90 |
ENSDART00000170293
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr6_+_56224631 | 1.89 |
ENSDART00000174612
ENSDART00000177824 |
FO834918.1
|
ENSDARG00000106015 |
| chr16_-_44706998 | 1.88 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr7_-_58856806 | 1.80 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr19_-_6215704 | 1.79 |
ENSDART00000104978
|
cica
|
capicua transcriptional repressor a |
| chr8_-_31374705 | 1.78 |
ENSDART00000162872
|
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr8_-_4704361 | 1.78 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr22_-_16351654 | 1.77 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr23_-_27579257 | 1.76 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr4_+_16020464 | 1.75 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr25_+_14773607 | 1.74 |
ENSDART00000171835
|
dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr14_-_15777250 | 1.73 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr1_+_22160932 | 1.70 |
ENSDART00000016488
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr19_+_1148544 | 1.70 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr4_+_13413303 | 1.69 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr4_-_71752866 | 1.68 |
ENSDART00000143417
ENSDART00000172042 |
zgc:162958
|
zgc:162958 |
| chr2_+_3115593 | 1.68 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr5_+_36168475 | 1.68 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr13_+_45387478 | 1.65 |
ENSDART00000019113
|
tmem57b
|
transmembrane protein 57b |
| chr5_+_32698516 | 1.65 |
ENSDART00000097945
|
usp20
|
ubiquitin specific peptidase 20 |
| chr24_+_23571680 | 1.65 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr6_+_35811398 | 1.64 |
ENSDART00000151760
|
FP236809.1
|
ENSDARG00000096356 |
| chr14_-_33605295 | 1.64 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr5_-_23291887 | 1.63 |
ENSDART00000099083
ENSDART00000099084 ENSDART00000147887 |
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr12_-_33558313 | 1.63 |
ENSDART00000111259
|
tmem94
|
transmembrane protein 94 |
| chr14_-_44880144 | 1.62 |
ENSDART00000163543
|
abhd18
|
abhydrolase domain containing 18 |
| chr9_+_29737843 | 1.60 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr21_+_347169 | 1.59 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr24_+_40770010 | 1.57 |
|
|
|
| chr13_+_45387708 | 1.56 |
ENSDART00000074567
|
tmem57b
|
transmembrane protein 57b |
| chr3_+_61903372 | 1.55 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr25_+_4414838 | 1.55 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr17_+_11519212 | 1.54 |
|
|
|
| chr13_+_22973830 | 1.54 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr10_-_25366450 | 1.54 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr6_+_38775771 | 1.53 |
ENSDART00000129655
|
ube3a
|
ubiquitin protein ligase E3A |
| chr20_+_34091702 | 1.53 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr17_+_49995001 | 1.51 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr5_+_23559435 | 1.50 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr21_-_14735450 | 1.50 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr15_-_20189604 | 1.49 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr23_-_14118530 | 1.49 |
ENSDART00000139406
|
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr17_-_49994799 | 1.48 |
|
|
|
| chr22_-_29957382 | 1.48 |
ENSDART00000019786
ENSDART00000159813 |
smc3
|
structural maintenance of chromosomes 3 |
| chr5_-_23171454 | 1.47 |
ENSDART00000135153
|
TBC1D8B
|
TBC1 domain family member 8B |
| chr7_-_58856862 | 1.46 |
ENSDART00000164104
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr8_-_22492972 | 1.45 |
|
|
|
| chr20_+_21491468 | 1.45 |
ENSDART00000049586
ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr6_+_41455676 | 1.44 |
ENSDART00000007798
ENSDART00000162135 |
twf2a
|
twinfilin actin-binding protein 2a |
| chr12_+_18776712 | 1.43 |
|
|
|
| chr7_-_25991876 | 1.43 |
ENSDART00000101109
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr11_+_24583090 | 1.42 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr24_+_23571827 | 1.42 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr20_-_47576872 | 1.41 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr11_+_5883229 | 1.38 |
ENSDART00000129663
|
dazap1
|
DAZ associated protein 1 |
| chr3_-_20643874 | 1.38 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr2_+_30800532 | 1.37 |
|
|
|
| chr25_+_18910528 | 1.36 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr8_-_11286308 | 1.35 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr15_-_20592699 | 1.35 |
ENSDART00000048423
|
timm50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr23_+_6652454 | 1.35 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr8_-_41511321 | 1.34 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr20_+_27813565 | 1.33 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr8_-_25826973 | 1.33 |
ENSDART00000047008
ENSDART00000128829 |
efhd2
|
EF-hand domain family, member D2 |
| chr25_-_1243081 | 1.32 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr8_-_37072857 | 1.32 |
ENSDART00000004041
|
zgc:162200
|
zgc:162200 |
| chr15_-_31266460 | 1.31 |
ENSDART00000157145
ENSDART00000155473 |
ksr1b
|
kinase suppressor of ras 1b |
| chr3_+_17901295 | 1.30 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr10_+_33629943 | 1.28 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr5_+_26537518 | 1.27 |
|
|
|
| chr20_-_46563521 | 1.27 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr1_+_33962885 | 1.26 |
ENSDART00000145535
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr2_+_30506215 | 1.25 |
ENSDART00000115271
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr7_+_71585147 | 1.24 |
ENSDART00000161344
|
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr7_-_37283707 | 1.24 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr4_-_20092548 | 1.24 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr16_+_9509875 | 1.24 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr19_-_31212648 | 1.23 |
ENSDART00000125893
ENSDART00000145581 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr8_-_18869085 | 1.22 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr14_-_33141111 | 1.21 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr7_-_25991743 | 1.21 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr13_+_46514282 | 1.21 |
ENSDART00000159260
|
CU695232.1
|
ENSDARG00000098286 |
| chr4_+_15007453 | 1.20 |
ENSDART00000146156
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr6_+_41506220 | 1.19 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr1_+_28379996 | 1.19 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr11_-_24300849 | 1.19 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr20_-_44678938 | 1.19 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr10_+_28273581 | 1.19 |
ENSDART00000131003
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr5_+_20418001 | 1.18 |
ENSDART00000131838
|
si:dkey-174n20.1
|
si:dkey-174n20.1 |
| chr16_+_50489655 | 1.18 |
|
|
|
| chr15_-_545279 | 1.18 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr8_-_37072691 | 1.18 |
ENSDART00000004041
|
zgc:162200
|
zgc:162200 |
| chr8_-_37072649 | 1.17 |
ENSDART00000004041
|
zgc:162200
|
zgc:162200 |
| chr11_-_10472636 | 1.17 |
ENSDART00000081827
|
ect2
|
epithelial cell transforming 2 |
| chr11_+_36088518 | 1.16 |
|
|
|
| chr5_-_19631924 | 1.13 |
ENSDART00000158613
|
usp30
|
ubiquitin specific peptidase 30 |
| chr20_+_1308632 | 1.13 |
ENSDART00000064436
|
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr7_-_9628384 | 1.13 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr19_+_10477101 | 1.13 |
ENSDART00000151735
|
necap1
|
NECAP endocytosis associated 1 |
| chr24_+_25872005 | 1.13 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr1_-_11191824 | 1.13 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr5_+_25408121 | 1.11 |
|
|
|
| chr8_-_48679386 | 1.11 |
ENSDART00000047134
|
clcn6
|
chloride channel 6 |
| chr6_+_13766964 | 1.11 |
ENSDART00000136006
ENSDART00000009382 |
dnpep
|
aspartyl aminopeptidase |
| chr22_+_17188103 | 1.11 |
ENSDART00000090107
|
nrd1b
|
nardilysin b (N-arginine dibasic convertase) |
| chr18_-_15300493 | 1.10 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr16_-_44707135 | 1.10 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr11_-_43409685 | 1.10 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr4_-_3340315 | 1.09 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr3_-_19890717 | 1.08 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr15_-_545179 | 1.08 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr5_-_32698092 | 1.08 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr13_+_16389663 | 1.08 |
|
|
|
| chr4_-_14916416 | 1.08 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr5_-_23291809 | 1.07 |
ENSDART00000099083
ENSDART00000099084 |
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr20_-_54285455 | 1.07 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr13_+_12538980 | 1.07 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr2_-_41888389 | 1.07 |
|
|
|
| chr13_-_42548129 | 1.06 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr6_+_33552518 | 1.06 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr23_-_36469779 | 1.06 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr6_+_46307828 | 1.05 |
ENSDART00000154817
|
si:dkeyp-67f1.1
|
si:dkeyp-67f1.1 |
| chr7_+_41607231 | 1.05 |
ENSDART00000165789
ENSDART00000115090 |
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr21_-_13026036 | 1.04 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 1.9 | 5.7 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.5 | 8.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 1.0 | 3.1 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.9 | 6.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.8 | 2.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.7 | 2.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.7 | 2.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.6 | 7.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.6 | 6.9 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.5 | 3.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 2.6 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.5 | 2.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.5 | 1.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 | 2.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.4 | 1.2 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.4 | 1.5 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.4 | 3.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 5.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.3 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 3.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.3 | 3.8 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.3 | 1.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.3 | 0.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 0.8 | GO:0071887 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.3 | 1.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 5.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 0.7 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.2 | 0.9 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.2 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.6 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.6 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 1.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 2.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.7 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 1.2 | GO:0045620 | negative regulation of lymphocyte differentiation(GO:0045620) negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 2.0 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.2 | 0.8 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.2 | 2.0 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 3.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 1.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.8 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 1.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.4 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 6.3 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 0.5 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 4.2 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 0.7 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.8 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 4.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 2.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 1.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.7 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.8 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.8 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 2.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.3 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 1.9 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 3.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.6 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.8 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 2.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.6 | GO:0009086 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.6 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 1.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.4 | GO:0033157 | regulation of intracellular protein transport(GO:0033157) positive regulation of intracellular protein transport(GO:0090316) |
| 0.0 | 1.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 2.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.9 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0051340 | regulation of ligase activity(GO:0051340) positive regulation of ligase activity(GO:0051351) |
| 0.0 | 1.7 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 1.0 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.3 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.1 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.5 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 1.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0000801 | central element(GO:0000801) |
| 0.8 | 9.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.6 | 6.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.5 | 6.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 2.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 4.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 1.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 1.8 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.3 | 1.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 6.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 4.1 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 6.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 6.0 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 2.5 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 4.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 10.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 2.3 | 6.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.9 | 7.7 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.7 | 5.0 | GO:0008425 | 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 1.1 | 6.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.8 | 6.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.7 | 2.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.7 | 2.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.6 | 3.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.6 | 2.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 2.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 1.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 3.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 1.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.4 | 6.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 1.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 2.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 0.9 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 0.8 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 0.8 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.3 | 0.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 2.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 4.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 6.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.6 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 1.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 3.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 6.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 3.9 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 2.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 8.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 7.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 4.3 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.8 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.5 | 3.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 1.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 4.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 6.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.1 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.5 | 1.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 3.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 5.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 7.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 4.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 3.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 5.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |