Project
DANIO-CODE
Navigation
Downloads
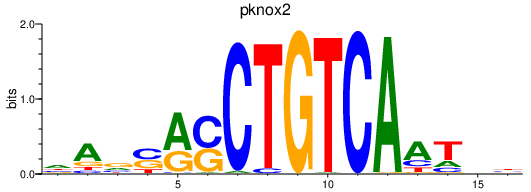
Results for pknox2
Z-value: 0.70
Transcription factors associated with pknox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pknox2
|
ENSDARG00000055349 | pbx/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pknox2 | dr10_dc_chr10_-_31334198_31334221 | 0.83 | 7.4e-05 | Click! |
Activity profile of pknox2 motif
Sorted Z-values of pknox2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pknox2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_14822990 | 2.42 |
ENSDART00000149949
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr20_+_28531684 | 2.27 |
ENSDART00000123387
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr18_+_29424354 | 2.17 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr6_+_14823185 | 2.12 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr11_+_27970922 | 1.75 |
ENSDART00000169360
ENSDART00000043756 |
ephb2b
|
eph receptor B2b |
| chr11_-_32461160 | 1.69 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr4_+_12932838 | 1.68 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr12_+_36796886 | 1.53 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr10_+_26020615 | 1.51 |
ENSDART00000128292
ENSDART00000170275 ENSDART00000166164 ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr21_-_34623741 | 1.50 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr2_+_3864843 | 1.47 |
ENSDART00000108964
ENSDART00000128514 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr2_-_55737505 | 1.35 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr12_+_17382681 | 1.26 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr3_-_36130248 | 1.20 |
ENSDART00000126588
|
rac3a
|
ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) |
| chr18_+_24575639 | 1.15 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr6_-_10593395 | 1.13 |
ENSDART00000020261
|
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr10_+_33926312 | 1.12 |
ENSDART00000174730
|
BX569785.2
|
ENSDARG00000106629 |
| chr22_-_23238714 | 1.04 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr4_+_12932950 | 1.02 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr21_+_17073163 | 0.90 |
|
|
|
| chr3_-_15739545 | 0.89 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr10_+_31701855 | 0.87 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr7_+_20215723 | 0.86 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr23_-_20402258 | 0.86 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr16_-_7321943 | 0.84 |
ENSDART00000149260
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr16_-_543957 | 0.83 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr4_-_16356071 | 0.81 |
ENSDART00000079521
|
kera
|
keratocan |
| chr19_+_41488385 | 0.78 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr5_+_41497442 | 0.77 |
ENSDART00000171919
ENSDART00000097587 |
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_-_10342940 | 0.76 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr5_+_15167637 | 0.74 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr17_-_21179064 | 0.72 |
|
|
|
| chr3_-_15739701 | 0.72 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr14_-_32553399 | 0.71 |
ENSDART00000169380
|
slc25a43
|
solute carrier family 25, member 43 |
| chr2_-_32575527 | 0.71 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr4_-_19027117 | 0.71 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr9_-_33518818 | 0.70 |
ENSDART00000140039
|
rpl8
|
ribosomal protein L8 |
| chr12_-_36565562 | 0.70 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr8_-_17148743 | 0.68 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr19_+_21783111 | 0.68 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr19_-_27730419 | 0.66 |
ENSDART00000151047
|
sh3bp5lb
|
SH3-binding domain protein 5-like, b |
| chr3_+_17387551 | 0.66 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr5_+_26537490 | 0.65 |
|
|
|
| chr3_-_15889742 | 0.62 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr5_-_40524018 | 0.59 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr12_+_49071673 | 0.57 |
|
|
|
| chr16_-_13704905 | 0.56 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr23_-_19177660 | 0.55 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr11_-_18004437 | 0.54 |
ENSDART00000125800
|
CABZ01112210.1
|
ENSDARG00000088766 |
| chr25_-_21796677 | 0.54 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr13_+_28286826 | 0.53 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr4_-_73683521 | 0.53 |
|
|
|
| chr1_-_13547500 | 0.53 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr16_+_40625705 | 0.53 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr2_-_41873625 | 0.52 |
ENSDART00000155577
|
ENSDARG00000056490
|
ENSDARG00000056490 |
| chr20_+_32575184 | 0.51 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr15_+_16961487 | 0.51 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr16_+_1073570 | 0.49 |
|
|
|
| chr21_-_38106145 | 0.48 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr19_+_21783429 | 0.47 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr14_+_50164568 | 0.47 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr9_-_30463616 | 0.42 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr12_-_21918058 | 0.41 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr25_+_3884499 | 0.41 |
ENSDART00000104926
|
ENSDARG00000058108
|
ENSDARG00000058108 |
| chr10_-_43200499 | 0.41 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr19_-_9793494 | 0.40 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr4_+_20901783 | 0.39 |
|
|
|
| chr19_+_1148444 | 0.39 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr11_-_18004011 | 0.38 |
ENSDART00000125800
|
CABZ01112210.1
|
ENSDARG00000088766 |
| chr23_+_34037058 | 0.37 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr4_-_3215227 | 0.37 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr13_-_28133363 | 0.36 |
ENSDART00000041036
|
smap1
|
small ArfGAP 1 |
| chr10_-_28474085 | 0.35 |
|
|
|
| chr6_+_14823900 | 0.33 |
ENSDART00000178555
|
CU929521.3
|
ENSDARG00000108533 |
| chr19_-_9793444 | 0.33 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr6_-_39162845 | 0.32 |
ENSDART00000148661
|
stat2
|
signal transducer and activator of transcription 2 |
| chr3_-_20972827 | 0.31 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr20_-_47576935 | 0.31 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr15_-_46484516 | 0.31 |
ENSDART00000154577
|
zgc:162872
|
zgc:162872 |
| chr19_+_41488351 | 0.30 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr23_+_18176720 | 0.30 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr7_-_68033217 | 0.28 |
|
|
|
| chr10_-_22988906 | 0.28 |
|
|
|
| chr2_+_26632673 | 0.28 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr19_+_43124105 | 0.27 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr5_+_26217533 | 0.27 |
|
|
|
| chr16_-_52761159 | 0.27 |
ENSDART00000111869
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_+_30416440 | 0.24 |
ENSDART00000012132
|
cbwd
|
COBW domain containing |
| chr11_-_27970634 | 0.24 |
|
|
|
| chr19_-_25843036 | 0.23 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr19_+_44310050 | 0.23 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr19_-_1921345 | 0.22 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr7_+_69664384 | 0.22 |
ENSDART00000058763
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr5_+_31889674 | 0.20 |
ENSDART00000165417
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr16_+_13964949 | 0.20 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr24_-_12836986 | 0.18 |
ENSDART00000133166
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr9_-_34410053 | 0.17 |
ENSDART00000111027
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr17_-_11264425 | 0.17 |
ENSDART00000151847
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr22_-_21021866 | 0.16 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr21_+_26502430 | 0.15 |
ENSDART00000021121
|
stx5al
|
syntaxin 5A, like |
| chr17_-_14772485 | 0.15 |
ENSDART00000080401
ENSDART00000154690 |
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr19_-_35739052 | 0.15 |
|
|
|
| chr23_-_4080767 | 0.14 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr20_-_47576610 | 0.14 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr5_-_34600498 | 0.13 |
ENSDART00000164396
|
fcho2
|
FCH domain only 2 |
| chr5_+_57073554 | 0.13 |
ENSDART00000166098
ENSDART00000005090 |
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr21_+_43887079 | 0.12 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr5_-_13145749 | 0.12 |
ENSDART00000166957
|
purba
|
purine-rich element binding protein Ba |
| chr12_-_17079132 | 0.12 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr10_+_10780578 | 0.11 |
ENSDART00000170147
ENSDART00000004181 |
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr7_-_56465497 | 0.11 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr6_+_49927433 | 0.11 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr16_+_3814595 | 0.11 |
ENSDART00000174521
|
CABZ01088996.1
|
ENSDARG00000108584 |
| chr19_-_35739013 | 0.11 |
|
|
|
| chr10_+_20632757 | 0.10 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_-_13266357 | 0.09 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr3_-_15739926 | 0.08 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr3_-_10343029 | 0.08 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr5_-_13267041 | 0.08 |
|
|
|
| chr1_+_19156170 | 0.07 |
ENSDART00000111454
|
ENSDARG00000080019
|
ENSDARG00000080019 |
| chr23_-_6442213 | 0.07 |
ENSDART00000112667
|
ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_663662 | 0.05 |
ENSDART00000125969
|
ube2ka
|
ubiquitin-conjugating enzyme E2Ka (UBC1 homolog, yeast) |
| chr7_-_29452646 | 0.05 |
ENSDART00000112651
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr18_-_33832 | 0.05 |
ENSDART00000158957
ENSDART00000129125 |
pde8a
|
phosphodiesterase 8A |
| chr19_-_6215704 | 0.04 |
ENSDART00000104978
|
cica
|
capicua transcriptional repressor a |
| chr9_-_33518171 | 0.04 |
ENSDART00000160534
ENSDART00000132973 |
rpl8
|
ribosomal protein L8 |
| chr24_-_17089437 | 0.00 |
|
|
|
| chr7_-_66641207 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 0.7 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.1 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 3.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.4 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 1.5 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 2.7 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.7 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.8 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.0 | 0.3 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.5 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.0 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 1.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.5 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.8 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.8 | GO:0004931 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |