Project
DANIO-CODE
Navigation
Downloads
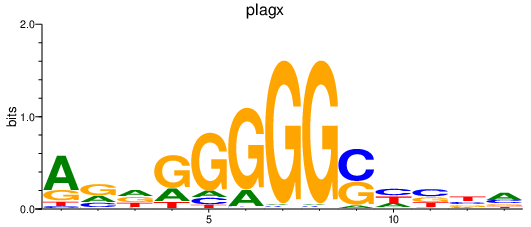
Results for plagx
Z-value: 0.72
Transcription factors associated with plagx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
plagx
|
ENSDARG00000036855 | pleiomorphic adenoma gene X |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| plagx | dr10_dc_chr23_+_32085181_32085304 | 0.77 | 4.8e-04 | Click! |
Activity profile of plagx motif
Sorted Z-values of plagx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of plagx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22499899 | 3.12 |
ENSDART00000147358
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr7_-_69639922 | 2.18 |
|
|
|
| chr20_-_44598129 | 2.15 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr15_+_43776204 | 2.05 |
ENSDART00000122238
|
tyr
|
tyrosinase |
| chr24_+_35676505 | 1.81 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr23_+_45020761 | 1.58 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr16_-_35633520 | 1.35 |
ENSDART00000169868
ENSDART00000166606 |
scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr12_-_25825072 | 1.31 |
|
|
|
| chr7_+_44269171 | 1.26 |
ENSDART00000149981
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr4_+_22020246 | 1.18 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr12_-_30230492 | 1.09 |
ENSDART00000152985
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr19_-_4079427 | 1.06 |
ENSDART00000170325
|
map7d1b
|
MAP7 domain containing 1b |
| chr5_+_63103958 | 1.01 |
|
|
|
| chr22_-_15930756 | 0.98 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr1_+_48923897 | 0.97 |
ENSDART00000101004
ENSDART00000164000 ENSDART00000142957 |
col17a1a
|
collagen, type XVII, alpha 1a |
| chr19_+_9113356 | 0.97 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr12_+_40919018 | 0.96 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr4_+_19545842 | 0.93 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr25_+_28329427 | 0.73 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr19_+_38033219 | 0.71 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr19_-_20508993 | 0.67 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr19_+_21783111 | 0.64 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr1_+_46754799 | 0.62 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr1_+_52373036 | 0.54 |
ENSDART00000129747
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr10_+_41237437 | 0.52 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr7_+_73594790 | 0.43 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr12_+_16391933 | 0.33 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr13_-_22653869 | 0.32 |
|
|
|
| chr3_-_25290558 | 0.32 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr3_+_18645970 | 0.27 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr24_-_35646129 | 0.27 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr2_-_24330667 | 0.27 |
|
|
|
| chr6_-_35417797 | 0.24 |
ENSDART00000016586
|
rgs5a
|
regulator of G protein signaling 5a |
| chr3_+_35377667 | 0.21 |
|
|
|
| chr21_-_20983913 | 0.20 |
ENSDART00000132091
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr16_-_36844763 | 0.19 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr16_+_35633745 | 0.10 |
|
|
|
| chr2_-_24330786 | 0.07 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.3 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 3.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 1.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 1.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.2 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 2.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |