Project
DANIO-CODE
Navigation
Downloads
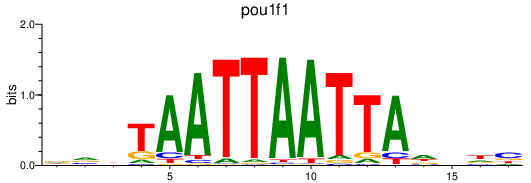
Results for pou1f1
Z-value: 1.38
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr10_dc_chr9_-_19154264_19154270 | 0.86 | 2.2e-05 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17590507 | 10.55 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr4_+_9668755 | 6.88 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr13_+_24148599 | 6.67 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr3_-_32686790 | 4.97 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr21_+_25199691 | 4.93 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr3_+_28808901 | 4.61 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr7_+_58718614 | 4.00 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr9_-_22402341 | 3.93 |
ENSDART00000110656
|
crygm2d20
|
crystallin, gamma M2d20 |
| chr20_+_19613133 | 3.68 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr23_+_19887319 | 3.57 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr23_-_42775849 | 3.48 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr23_-_6588073 | 3.15 |
ENSDART00000092214
ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr14_+_47326080 | 3.12 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr15_+_9351511 | 3.07 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr10_+_18994733 | 3.03 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr20_-_9107294 | 2.99 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr22_-_22846670 | 2.91 |
ENSDART00000176355
|
CABZ01039424.2
|
ENSDARG00000108442 |
| chr24_-_1155215 | 2.89 |
ENSDART00000177356
|
itgb1a
|
integrin, beta 1a |
| chr7_-_73866202 | 2.81 |
|
|
|
| chr16_+_46145286 | 2.74 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr24_+_32261533 | 2.69 |
ENSDART00000003745
|
vim
|
vimentin |
| chr19_+_43763483 | 2.68 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr11_+_30416115 | 2.66 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr15_+_43714699 | 2.53 |
ENSDART00000168191
|
ENSDARG00000102235
|
ENSDARG00000102235 |
| chr19_+_44185325 | 2.50 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr21_-_43020159 | 2.47 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr6_-_16590883 | 2.46 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr3_-_15849644 | 2.45 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_+_40722565 | 2.31 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr16_+_43249142 | 2.28 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr10_+_9575 | 2.26 |
|
|
|
| chr19_-_39048324 | 2.24 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr4_+_4841191 | 2.23 |
ENSDART00000130818
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr9_+_36092350 | 2.22 |
ENSDART00000005086
|
atp1a1b
|
ATPase, Na+/K+ transporting, alpha 1b polypeptide |
| chr5_-_40894631 | 2.19 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr9_-_1949904 | 2.14 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr16_-_28921444 | 2.12 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr4_-_73485204 | 2.12 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr23_-_21520413 | 2.11 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr16_+_46435014 | 2.08 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr2_+_20748431 | 2.08 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr19_-_35739239 | 2.04 |
|
|
|
| chr14_-_24094142 | 2.00 |
ENSDART00000126894
|
fam13b
|
family with sequence similarity 13, member B |
| chr17_-_30685367 | 1.99 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr14_-_4038642 | 1.97 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr19_+_12995955 | 1.97 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr1_-_53403731 | 1.93 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr7_+_19243998 | 1.89 |
ENSDART00000173766
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr14_-_45883839 | 1.86 |
|
|
|
| chr21_+_45802046 | 1.85 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr7_+_25758469 | 1.80 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr1_+_4704661 | 1.78 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr5_+_62987426 | 1.78 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr19_-_27284726 | 1.76 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr4_+_1750689 | 1.76 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr2_+_16492315 | 1.75 |
ENSDART00000135783
|
selj
|
selenoprotein J |
| chr12_+_2988101 | 1.73 |
ENSDART00000044690
ENSDART00000122905 |
rac3b
|
ras-related C3 botulinum toxin substrate 3b (rho family, small GTP binding protein Rac3) |
| chr5_-_44229464 | 1.73 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr16_-_28943421 | 1.71 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr13_-_49983998 | 1.70 |
|
|
|
| chr11_-_1374026 | 1.68 |
ENSDART00000172953
|
rpl29
|
ribosomal protein L29 |
| chr5_+_71028018 | 1.67 |
ENSDART00000164893
ENSDART00000159658 ENSDART00000097164 ENSDART00000124939 ENSDART00000171230 |
LHX3
|
LIM homeobox 3 |
| chr1_-_22160662 | 1.66 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr4_+_22759177 | 1.65 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr2_+_29729161 | 1.65 |
ENSDART00000141666
|
AL845362.1
|
ENSDARG00000093205 |
| chr13_-_9554501 | 1.64 |
ENSDART00000139541
|
ENSDARG00000069828
|
ENSDARG00000069828 |
| chr13_+_9100463 | 1.63 |
ENSDART00000058064
|
ENSDARG00000039726
|
ENSDARG00000039726 |
| chr15_-_18226612 | 1.62 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr24_+_9335428 | 1.60 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr14_+_2969679 | 1.57 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr20_+_7596461 | 1.56 |
ENSDART00000127975
ENSDART00000132481 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr4_-_2615160 | 1.55 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr14_-_16791447 | 1.55 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr5_-_9162684 | 1.54 |
|
|
|
| chr10_-_43924675 | 1.54 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr13_+_7109810 | 1.53 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr18_+_24932972 | 1.53 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr21_+_28441951 | 1.53 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr3_+_50511676 | 1.52 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr12_+_27040886 | 1.51 |
|
|
|
| chr5_-_2014043 | 1.48 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr2_-_44402486 | 1.46 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr9_+_34832049 | 1.46 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr15_-_21003820 | 1.44 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr4_-_935448 | 1.42 |
ENSDART00000171855
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr12_+_13244482 | 1.42 |
ENSDART00000137757
|
irf9
|
interferon regulatory factor 9 |
| chr12_+_13870214 | 1.41 |
ENSDART00000066367
|
fkbp10b
|
FK506 binding protein 10b |
| chr21_+_44203036 | 1.41 |
|
|
|
| chr6_-_50705420 | 1.41 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr10_+_13042496 | 1.40 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr20_-_3970778 | 1.40 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr3_-_19050721 | 1.39 |
ENSDART00000131503
|
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr24_+_24686591 | 1.39 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr8_+_50994889 | 1.39 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr11_-_26595578 | 1.38 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr10_-_35293024 | 1.38 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr6_+_52350428 | 1.36 |
ENSDART00000151612
|
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr18_+_21419735 | 1.36 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr20_+_3095763 | 1.35 |
ENSDART00000133435
|
CEP170B
|
centrosomal protein 170B |
| chr21_-_30508374 | 1.34 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr19_+_20181908 | 1.33 |
ENSDART00000158987
|
hoxa4a
|
homeobox A4a |
| chr19_+_5562107 | 1.33 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr7_-_73866265 | 1.33 |
|
|
|
| chr4_+_5246465 | 1.33 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr8_-_17031599 | 1.33 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr13_-_31491759 | 1.32 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr14_-_33483075 | 1.32 |
ENSDART00000158870
|
si:dkey-76i15.1
|
si:dkey-76i15.1 |
| chr11_+_15935769 | 1.30 |
ENSDART00000158824
|
ctbs
|
chitobiase, di-N-acetyl- |
| chr1_+_39844690 | 1.29 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr24_+_37692365 | 1.29 |
|
|
|
| chr14_-_47129958 | 1.29 |
ENSDART00000172166
|
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr10_+_26786051 | 1.26 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr17_+_25500291 | 1.24 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr24_-_24306469 | 1.21 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr8_+_2428689 | 1.20 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr21_+_6858073 | 1.18 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr7_+_21006803 | 1.16 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr15_+_9351211 | 1.14 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr8_+_24725855 | 1.14 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr17_-_36948926 | 1.13 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr6_+_6640324 | 1.13 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr8_+_43334181 | 1.12 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr1_+_40795366 | 1.11 |
ENSDART00000146310
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr16_-_2570425 | 1.10 |
|
|
|
| chr2_-_17721575 | 1.10 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr16_+_9141325 | 1.06 |
|
|
|
| chr2_+_29507819 | 1.05 |
|
|
|
| chr10_-_35714800 | 1.05 |
|
|
|
| chr14_-_47083485 | 1.05 |
ENSDART00000172512
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr25_-_21718782 | 1.04 |
ENSDART00000175132
|
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr5_-_40894693 | 1.03 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr2_+_21267793 | 1.03 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr14_-_31514534 | 1.03 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr8_+_28240085 | 1.01 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr10_-_11303185 | 1.01 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr8_-_17951834 | 1.00 |
ENSDART00000112699
|
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr2_+_44718147 | 0.99 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr2_-_44402452 | 0.98 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr5_+_65754237 | 0.98 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr7_+_66660781 | 0.97 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr14_-_30630835 | 0.97 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr3_-_12087518 | 0.97 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr8_-_12253026 | 0.97 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr16_+_43249077 | 0.97 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr16_+_53602834 | 0.96 |
ENSDART00000074653
|
grinab
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1b (glutamate binding) |
| chr24_+_192889 | 0.96 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr20_+_36098607 | 0.96 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr13_+_33138144 | 0.95 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr1_-_30801520 | 0.94 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr16_+_37519702 | 0.94 |
|
|
|
| chr1_-_661529 | 0.93 |
ENSDART00000166786
ENSDART00000170483 |
appa
|
amyloid beta (A4) precursor protein a |
| chr2_+_4144790 | 0.92 |
ENSDART00000160806
|
mkxb
|
mohawk homeobox b |
| chr15_-_21078596 | 0.91 |
ENSDART00000154019
|
ENSDARG00000093199
|
ENSDARG00000093199 |
| chr12_-_41128584 | 0.90 |
|
|
|
| chr21_+_27241416 | 0.90 |
|
|
|
| chr12_-_28248133 | 0.90 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr4_-_885413 | 0.90 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr15_+_32439470 | 0.90 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr20_+_25326042 | 0.89 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr16_+_29060022 | 0.88 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr9_-_43736549 | 0.87 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr23_-_23474703 | 0.85 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr9_+_7745593 | 0.85 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr6_-_51666164 | 0.85 |
|
|
|
| chr12_-_425381 | 0.84 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr5_-_55263153 | 0.84 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr17_+_2551397 | 0.84 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr9_+_24255064 | 0.83 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr8_-_2375664 | 0.83 |
ENSDART00000144017
|
vdac3
|
voltage-dependent anion channel 3 |
| chr13_-_29290894 | 0.83 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr19_-_39048402 | 0.83 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr14_-_28228583 | 0.83 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr3_-_19218660 | 0.82 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr12_-_552235 | 0.81 |
ENSDART00000168586
ENSDART00000158355 |
bsk146
|
brain specific kinase 146 |
| chr18_-_48375325 | 0.80 |
|
|
|
| chr16_+_21065200 | 0.77 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr18_-_43890514 | 0.77 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr8_-_20357907 | 0.76 |
|
|
|
| chr6_+_9192009 | 0.76 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr25_-_25339268 | 0.75 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr6_+_14854101 | 0.75 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr17_+_10345498 | 0.73 |
ENSDART00000081659
ENSDART00000169120 ENSDART00000170970 ENSDART00000140391 |
tyro3
|
TYRO3 protein tyrosine kinase |
| chr7_-_29003965 | 0.73 |
|
|
|
| chr16_-_21334269 | 0.73 |
ENSDART00000145837
|
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr9_-_53380782 | 0.72 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr17_+_44583892 | 0.72 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr5_-_42304154 | 0.71 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr1_+_51087450 | 0.71 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr6_+_7376428 | 0.71 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr19_-_5851328 | 0.70 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr9_+_11311198 | 0.70 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr14_-_1342450 | 0.69 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr3_-_7220340 | 0.69 |
|
|
|
| chr3_-_38642067 | 0.69 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr18_-_13234871 | 0.68 |
ENSDART00000142942
|
klhl36
|
kelch-like family member 36 |
| chr7_-_71290605 | 0.67 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr8_-_20357968 | 0.67 |
|
|
|
| chr16_-_53008678 | 0.65 |
|
|
|
| chr12_-_22438379 | 0.65 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr14_-_23784679 | 0.65 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.8 | 2.5 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.8 | 2.4 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.5 | 1.6 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.5 | 1.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.5 | 5.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.5 | 1.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.5 | 1.8 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 1.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 1.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.4 | 2.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 0.9 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.3 | 0.8 | GO:0051701 | interaction with host(GO:0051701) |
| 0.3 | 1.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.3 | 1.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 1.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.3 | 0.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.3 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 1.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 1.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 2.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 1.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 2.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 1.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.2 | 1.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 1.3 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.2 | 2.9 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.2 | 0.8 | GO:0008291 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 1.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.6 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 2.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 4.0 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.4 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 2.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.7 | GO:0060753 | mast cell chemotaxis(GO:0002551) positive regulation of leukocyte chemotaxis(GO:0002690) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.7 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 0.3 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 6.7 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 0.6 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.6 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 3.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.0 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 7.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.7 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.6 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 2.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 3.3 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 4.2 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 0.1 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.0 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0046958 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 1.3 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 2.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.7 | GO:0042060 | wound healing(GO:0042060) |
| 0.0 | 0.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 2.9 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 1.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 6.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 1.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 1.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 2.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 4.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 7.3 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 1.2 | 4.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.0 | 2.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 5.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.5 | 4.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 3.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 0.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.3 | 2.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 7.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.9 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 3.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 3.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.2 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 1.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.6 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 2.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 2.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.1 | 1.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 18.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.5 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 1.2 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 1.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 2.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 4.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |