Project
DANIO-CODE
Navigation
Downloads
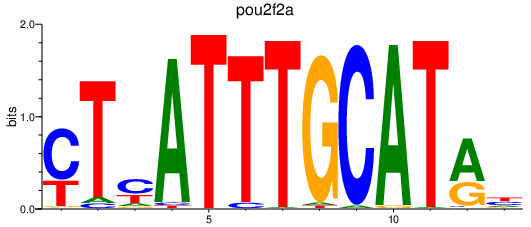
Results for pou2f2a
Z-value: 1.90
Transcription factors associated with pou2f2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f2a
|
ENSDARG00000019658 | POU class 2 homeobox 2a |
|
pou2f2a
|
ENSDARG00000036816 | POU class 2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f2a | dr10_dc_chr16_+_11027394_11027442 | 0.57 | 2.2e-02 | Click! |
Activity profile of pou2f2a motif
Sorted Z-values of pou2f2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_6078015 | 6.91 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr16_+_17808623 | 5.01 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr23_+_21546553 | 4.73 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr23_+_23305483 | 4.53 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr6_-_39315024 | 4.44 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr5_-_42272455 | 4.35 |
ENSDART00000161586
|
flot2a
|
flotillin 2a |
| chr6_+_56163589 | 3.87 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_+_737615 | 3.67 |
ENSDART00000161774
|
irx1a
|
iroquois homeobox 1a |
| chr15_-_29454583 | 3.62 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr9_-_43178744 | 3.45 |
|
|
|
| chr15_-_16076874 | 3.35 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr15_+_7190427 | 3.34 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr14_-_32624823 | 3.33 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr14_+_35885903 | 3.19 |
ENSDART00000052569
|
pitx2
|
paired-like homeodomain 2 |
| chr23_-_31446156 | 3.16 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr5_+_12786897 | 3.13 |
|
|
|
| chr20_+_40247918 | 3.04 |
ENSDART00000121818
|
trdn
|
triadin |
| chr5_-_65349550 | 2.99 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr10_+_17494274 | 2.97 |
|
|
|
| chr8_-_22945616 | 2.91 |
|
|
|
| chr3_+_28450576 | 2.89 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr9_-_54126121 | 2.87 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr16_-_24634751 | 2.84 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr13_-_5953330 | 2.84 |
ENSDART00000099224
|
dld
|
deltaD |
| chr11_-_11591394 | 2.80 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr20_-_22576513 | 2.74 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr11_-_29910947 | 2.59 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr8_+_28584427 | 2.56 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr14_-_40454194 | 2.52 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_53353690 | 2.52 |
ENSDART00000126339
|
dla
|
deltaA |
| chr17_+_26946957 | 2.51 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr1_-_46141359 | 2.51 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr19_+_22478256 | 2.47 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr6_-_343209 | 2.46 |
|
|
|
| chr23_-_4990023 | 2.45 |
ENSDART00000142699
|
taz
|
tafazzin |
| chr19_+_3713027 | 2.45 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_+_50395721 | 2.45 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr9_-_53898458 | 2.41 |
ENSDART00000175619
ENSDART00000179009 |
CABZ01078244.2
|
ENSDARG00000107473 |
| chr25_-_3704591 | 2.38 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr14_-_4038642 | 2.36 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr7_-_11812634 | 2.28 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr10_+_36096599 | 2.27 |
|
|
|
| chr14_-_7774262 | 2.27 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr9_-_53898524 | 2.27 |
ENSDART00000175619
ENSDART00000179009 |
CABZ01078244.2
|
ENSDARG00000107473 |
| chr17_+_33766838 | 2.24 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr25_-_2485276 | 2.24 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr1_-_24747855 | 2.21 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr10_+_10428297 | 2.18 |
ENSDART00000179214
|
sardh
|
sarcosine dehydrogenase |
| chr22_+_36687668 | 2.15 |
ENSDART00000134031
|
b3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr7_-_55406347 | 2.13 |
ENSDART00000021009
|
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr21_+_19034242 | 2.09 |
ENSDART00000123309
|
nkx6.1
|
NK6 homeobox 1 |
| chr2_+_55859099 | 2.08 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr11_+_5745644 | 2.07 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr24_-_8590572 | 2.06 |
ENSDART00000082349
|
tfap2a
|
transcription factor AP-2 alpha |
| chr11_-_25962291 | 2.05 |
|
|
|
| chr9_+_54612545 | 2.04 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr7_+_35769973 | 2.02 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr2_-_42784609 | 2.01 |
|
|
|
| chr22_-_336857 | 1.97 |
|
|
|
| chr7_-_33678058 | 1.95 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr12_+_23302859 | 1.94 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr15_-_18638643 | 1.93 |
ENSDART00000142010
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr10_+_4987494 | 1.92 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr6_-_14821305 | 1.92 |
|
|
|
| chr21_+_27346176 | 1.91 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr23_+_10552781 | 1.90 |
|
|
|
| chr5_-_32687349 | 1.89 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr15_+_1187249 | 1.88 |
ENSDART00000152638
ENSDART00000152466 |
mlf1
|
myeloid leukemia factor 1 |
| chr23_+_11412329 | 1.88 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr1_+_30992553 | 1.88 |
ENSDART00000112333
|
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr16_+_26989962 | 1.87 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr23_-_20332076 | 1.86 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr12_-_3742062 | 1.85 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr6_+_30441419 | 1.79 |
|
|
|
| chr14_-_27703812 | 1.77 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr5_+_44722544 | 1.74 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr14_-_26406720 | 1.74 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr7_+_65532390 | 1.74 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr17_-_26849495 | 1.71 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr23_-_1008307 | 1.70 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr5_-_24568752 | 1.70 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr13_-_37394157 | 1.69 |
ENSDART00000141420
|
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr23_-_29138952 | 1.68 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr1_+_45503061 | 1.68 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr25_+_18467217 | 1.67 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr11_+_27970922 | 1.64 |
ENSDART00000169360
ENSDART00000043756 |
ephb2b
|
eph receptor B2b |
| chr5_-_41672394 | 1.63 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr1_-_51086211 | 1.62 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr13_+_18414806 | 1.61 |
ENSDART00000110197
|
zgc:154058
|
zgc:154058 |
| chr13_+_24703802 | 1.61 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr9_-_55204516 | 1.60 |
|
|
|
| chr4_-_9779763 | 1.60 |
ENSDART00000134280
|
svopl
|
SVOP-like |
| chr19_+_20202737 | 1.54 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr18_+_38768524 | 1.53 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr10_+_10393377 | 1.47 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr2_+_38057483 | 1.43 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr12_-_36138709 | 1.42 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr21_+_38770125 | 1.41 |
ENSDART00000149085
|
hnf1bb
|
HNF1 homeobox Bb |
| chr1_+_12076622 | 1.40 |
ENSDART00000159226
|
pcdh10a
|
protocadherin 10a |
| chr25_+_5922190 | 1.39 |
ENSDART00000128816
|
sv2
|
synaptic vesicle glycoprotein 2 |
| chr11_+_24720057 | 1.37 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr8_-_38308535 | 1.37 |
|
|
|
| chr10_+_36861503 | 1.37 |
ENSDART00000153914
|
BX323076.1
|
ENSDARG00000097240 |
| chr17_+_18097490 | 1.35 |
ENSDART00000144894
|
bcl11ba
|
B-cell CLL/lymphoma 11Ba (zinc finger protein) |
| chr24_-_25283218 | 1.34 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_+_72257899 | 1.31 |
|
|
|
| chr19_-_28273791 | 1.31 |
ENSDART00000137033
|
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr19_+_20183210 | 1.31 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr12_-_34786844 | 1.30 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr22_-_337198 | 1.30 |
|
|
|
| chr25_+_5162045 | 1.29 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr21_+_5887486 | 1.27 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr13_+_41998500 | 1.26 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_-_54103765 | 1.26 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr2_-_30071872 | 1.25 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| KN150001v1_+_14939 | 1.22 |
|
|
|
| chr12_+_30471689 | 1.21 |
ENSDART00000124920
|
nrap
|
nebulin-related anchoring protein |
| chr3_-_58488929 | 1.21 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr19_-_22182031 | 1.20 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr19_+_14669633 | 1.19 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr2_+_33399405 | 1.19 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr5_-_57016269 | 1.19 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr12_-_28766713 | 1.18 |
ENSDART00000148459
|
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr16_+_53111969 | 1.17 |
ENSDART00000011506
|
nkd2b
|
naked cuticle homolog 2b |
| chr3_+_24145201 | 1.15 |
ENSDART00000169765
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr20_+_51388214 | 1.15 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr3_-_5318289 | 1.15 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr20_+_28958586 | 1.13 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr8_+_11604334 | 1.13 |
ENSDART00000004288
|
ift81
|
intraflagellar transport 81 homolog |
| chr16_+_12740993 | 1.13 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr23_-_4175790 | 1.10 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr21_+_5044859 | 1.10 |
ENSDART00000130569
|
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr11_+_19894772 | 1.10 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr21_+_44018278 | 1.09 |
ENSDART00000012396
|
ik
|
IK cytokine |
| chr19_-_30975279 | 1.08 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr12_-_3741925 | 1.08 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr5_-_62609495 | 1.08 |
ENSDART00000164651
|
gsnb
|
gelsolin b |
| chr4_-_16417703 | 1.08 |
ENSDART00000013085
|
dcn
|
decorin |
| chr5_+_58824344 | 1.07 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr25_+_34135377 | 1.07 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr2_+_10497066 | 1.06 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr15_-_47696481 | 1.05 |
|
|
|
| chr16_-_20084451 | 1.05 |
ENSDART00000079155
|
hdac9b
|
histone deacetylase 9b |
| chr5_-_24806610 | 1.04 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr19_+_42491631 | 1.04 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr1_+_41523935 | 1.03 |
ENSDART00000110860
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr5_-_34393017 | 1.02 |
ENSDART00000134516
|
btf3
|
basic transcription factor 3 |
| chr8_-_17031599 | 0.99 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr22_-_336942 | 0.99 |
|
|
|
| chr1_+_6995893 | 0.99 |
ENSDART00000163488
|
en1b
|
engrailed homeobox 1b |
| chr1_-_1254253 | 0.99 |
ENSDART00000130697
|
runx1
|
runt-related transcription factor 1 |
| chr1_-_47080507 | 0.98 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr20_+_51388313 | 0.97 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr17_+_1986151 | 0.95 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr5_-_24365811 | 0.95 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr11_+_19894390 | 0.93 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr2_+_58658615 | 0.93 |
|
|
|
| chr24_+_16992420 | 0.93 |
|
|
|
| chr5_+_63103958 | 0.92 |
|
|
|
| chr16_-_21064300 | 0.92 |
ENSDART00000040727
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr21_-_11554253 | 0.92 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr25_-_35698109 | 0.92 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3b |
| chr7_-_29300402 | 0.90 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr21_+_26696853 | 0.88 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr6_+_4379989 | 0.87 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr21_-_14276856 | 0.87 |
|
|
|
| chr2_-_30071815 | 0.86 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr16_+_25158949 | 0.84 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr24_+_16249188 | 0.83 |
ENSDART00000164516
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr16_+_21090083 | 0.82 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr23_-_6831711 | 0.82 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr4_+_67023074 | 0.81 |
ENSDART00000158749
|
zgc:173702
|
zgc:173702 |
| chr2_-_30071993 | 0.81 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr4_+_830826 | 0.80 |
|
|
|
| chr4_+_9393501 | 0.80 |
ENSDART00000092013
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr11_-_35313757 | 0.80 |
ENSDART00000031441
|
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr7_-_29300448 | 0.80 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr20_-_33272014 | 0.80 |
ENSDART00000178752
ENSDART00000047834 |
nbas
|
neuroblastoma amplified sequence |
| chr7_-_51186389 | 0.78 |
ENSDART00000174328
|
arhgap6
|
Rho GTPase activating protein 6 |
| KN150001v1_+_15017 | 0.77 |
|
|
|
| chr19_-_32454747 | 0.77 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr13_+_30820707 | 0.77 |
ENSDART00000057469
|
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr13_+_49154065 | 0.77 |
|
|
|
| chr3_-_45420882 | 0.77 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr23_-_2957203 | 0.76 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr19_+_42491275 | 0.76 |
|
|
|
| chr13_-_48465147 | 0.75 |
ENSDART00000132895
|
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr12_-_31342432 | 0.75 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr20_+_46309812 | 0.74 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr23_+_16937788 | 0.72 |
ENSDART00000060181
|
zgc:114174
|
zgc:114174 |
| chr20_+_25727285 | 0.72 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr2_-_59019569 | 0.72 |
|
|
|
| chr7_-_67028394 | 0.71 |
ENSDART00000048104
|
znf143a
|
zinc finger protein 143a |
| chr3_+_41416821 | 0.70 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr2_+_52561735 | 0.70 |
ENSDART00000175043
|
CABZ01040021.1
|
ENSDARG00000107973 |
| chr24_+_25326286 | 0.70 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| KN150460v1_+_164589 | 0.69 |
|
|
|
| chr2_+_47864913 | 0.69 |
|
|
|
| chr13_+_48565310 | 0.69 |
|
|
|
| chr2_-_42784707 | 0.68 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.8 | 2.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.8 | 3.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.8 | 3.1 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.6 | 2.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.6 | 3.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.6 | 2.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.6 | 3.3 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) foregut morphogenesis(GO:0007440) |
| 0.5 | 2.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.5 | 2.1 | GO:1901389 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.5 | 3.2 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.5 | 4.6 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.5 | 2.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 4.3 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.5 | 1.4 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.4 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 5.9 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.3 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 1.0 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of granulocyte differentiation(GO:0030852) plasminogen activation(GO:0031639) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of neutrophil differentiation(GO:0045658) |
| 0.3 | 3.9 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.3 | 2.9 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.3 | 2.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 2.8 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.3 | 1.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 2.5 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.3 | 1.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.3 | 1.2 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.3 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 0.8 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.7 | GO:1903959 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.5 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.2 | 2.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 3.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 2.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 1.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.2 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.7 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.2 | 3.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 3.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.7 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.2 | 2.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 2.0 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.1 | 1.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 1.1 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.3 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.8 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 2.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.5 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 6.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 2.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 2.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.5 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.1 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.0 | 1.1 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.6 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 5.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.7 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.8 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 4.3 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.8 | 2.4 | GO:0098594 | mucin granule(GO:0098594) |
| 0.6 | 1.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 4.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 3.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.9 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 7.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.8 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.6 | 2.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.6 | 3.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 1.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 1.3 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 2.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 1.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 5.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 2.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.3 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 5.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.7 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.2 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.2 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 3.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 3.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 38.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 5.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 1.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 4.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 2.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 2.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 4.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.1 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.0 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |