Project
DANIO-CODE
Navigation
Downloads
Results for pou2f3
Z-value: 0.54
Transcription factors associated with pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f3
|
ENSDARG00000052387 | POU class 2 homeobox 3 |
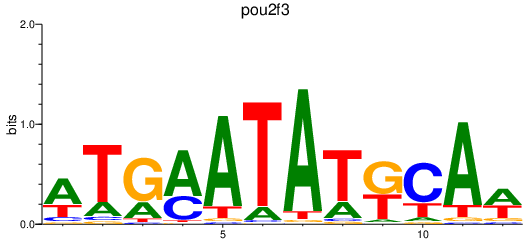
Activity profile of pou2f3 motif
Sorted Z-values of pou2f3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_13118866 | 0.84 |
ENSDART00000123257
|
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr8_-_14014576 | 0.76 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr21_+_36938230 | 0.58 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr10_-_22881453 | 0.57 |
|
|
|
| chr14_+_14850200 | 0.53 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr9_+_34510091 | 0.53 |
ENSDART00000035522
ENSDART00000170615 ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr9_+_49961721 | 0.46 |
|
|
|
| chr10_+_26636293 | 0.44 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr3_-_59690168 | 0.42 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_+_54525614 | 0.41 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr16_+_4292625 | 0.41 |
|
|
|
| chr10_-_45535333 | 0.39 |
|
|
|
| chr19_-_5186692 | 0.38 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr13_-_2211438 | 0.38 |
ENSDART00000137124
|
arfgef3
|
ARFGEF family member 3 |
| chr11_+_30416953 | 0.37 |
|
|
|
| chr16_+_46145286 | 0.37 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_-_5429428 | 0.36 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr4_+_30530 | 0.35 |
ENSDART00000157825
|
syn3
|
synapsin III |
| chr10_+_2554651 | 0.34 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr18_+_5303006 | 0.34 |
|
|
|
| chr3_-_48458042 | 0.33 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr10_+_15450466 | 0.33 |
ENSDART00000003839
|
nln
|
neurolysin (metallopeptidase M3 family) |
| chr8_-_7584890 | 0.33 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr9_+_30297924 | 0.32 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr10_-_14971401 | 0.32 |
ENSDART00000044756
ENSDART00000128579 |
smad2
|
SMAD family member 2 |
| chr13_+_22346304 | 0.32 |
ENSDART00000137220
|
ldb3a
|
LIM domain binding 3a |
| chr1_-_50395003 | 0.30 |
ENSDART00000035150
|
spast
|
spastin |
| chr5_-_47523737 | 0.30 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr20_-_43853614 | 0.29 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr5_-_19459698 | 0.29 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr5_-_15921657 | 0.29 |
|
|
|
| chr18_+_31038010 | 0.28 |
ENSDART00000163471
|
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr20_+_54525159 | 0.28 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr7_-_17916122 | 0.28 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr21_+_34053590 | 0.28 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr14_-_3937426 | 0.27 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr11_+_11000566 | 0.27 |
ENSDART00000065933
|
itgb6
|
integrin, beta 6 |
| chr19_+_1743359 | 0.27 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr24_-_20496410 | 0.26 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr9_+_12473243 | 0.26 |
ENSDART00000132709
|
tmem41aa
|
transmembrane protein 41aa |
| chr5_+_23741791 | 0.26 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr11_+_43136945 | 0.26 |
|
|
|
| chr18_-_20880226 | 0.25 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr20_-_54669179 | 0.25 |
|
|
|
| chr6_+_4091120 | 0.25 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr10_-_22948684 | 0.25 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr16_-_12282596 | 0.25 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr10_-_34058331 | 0.24 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr16_+_2582656 | 0.24 |
ENSDART00000092299
|
ENSDARG00000073784
|
ENSDARG00000073784 |
| chr10_+_9762039 | 0.24 |
|
|
|
| chr13_-_39033849 | 0.24 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr25_-_2932820 | 0.24 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr25_-_24960874 | 0.24 |
ENSDART00000178891
|
ENSDARG00000061421
|
ENSDARG00000061421 |
| chr22_+_35299551 | 0.23 |
ENSDART00000165353
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr16_-_43107682 | 0.23 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr3_+_18645970 | 0.23 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr16_-_33637966 | 0.22 |
ENSDART00000142965
|
BX511161.1
|
ENSDARG00000092272 |
| chr5_-_64983812 | 0.22 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr20_+_54595334 | 0.22 |
|
|
|
| chr8_+_22910147 | 0.22 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr1_+_51087450 | 0.22 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr10_+_2554914 | 0.22 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr5_-_34364552 | 0.22 |
ENSDART00000133170
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr10_-_36849596 | 0.22 |
|
|
|
| chr11_-_37613237 | 0.22 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr5_-_23741221 | 0.22 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr22_+_18364282 | 0.21 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr23_-_18131105 | 0.21 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr19_+_2942092 | 0.21 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr7_+_10792973 | 0.21 |
|
|
|
| chr18_+_3482740 | 0.21 |
ENSDART00000158763
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr19_+_2942485 | 0.21 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr21_-_40694583 | 0.21 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr18_+_1347470 | 0.21 |
|
|
|
| chr3_+_39425125 | 0.21 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr13_+_42276937 | 0.21 |
|
|
|
| chr8_-_53584884 | 0.20 |
|
|
|
| chr24_-_25283218 | 0.20 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr17_-_37447869 | 0.20 |
ENSDART00000148160
|
crip1
|
cysteine-rich protein 1 |
| chr9_+_3416525 | 0.20 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr23_-_33783345 | 0.20 |
ENSDART00000143333
|
pou6f1
|
POU class 6 homeobox 1 |
| chr19_+_2942373 | 0.20 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr20_-_39886349 | 0.20 |
ENSDART00000098253
|
rnf217
|
ring finger protein 217 |
| chr23_+_21546553 | 0.20 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr14_-_43191278 | 0.19 |
ENSDART00000169615
|
adh5
|
alcohol dehydrogenase 5 |
| chr21_+_26035166 | 0.19 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr25_-_36678227 | 0.19 |
ENSDART00000087247
|
glg1a
|
golgi glycoprotein 1a |
| chr23_-_27418761 | 0.19 |
ENSDART00000022042
|
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr10_+_23090883 | 0.19 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr11_-_41798903 | 0.19 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr10_-_42302932 | 0.19 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr24_-_32864493 | 0.19 |
|
|
|
| chr13_+_25590508 | 0.19 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr17_-_29196678 | 0.18 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr17_-_30958965 | 0.18 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr11_+_30400284 | 0.18 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr11_+_36147478 | 0.18 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr1_-_18581114 | 0.18 |
ENSDART00000178278
|
BX546494.1
|
ENSDARG00000107606 |
| chr4_-_1952230 | 0.18 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr20_-_1248089 | 0.18 |
ENSDART00000027119
|
lats1
|
large tumor suppressor kinase 1 |
| chr15_-_12297979 | 0.18 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr23_-_25871518 | 0.17 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr1_-_9256864 | 0.17 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr2_+_35258438 | 0.17 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr5_-_18393705 | 0.17 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr12_-_26317480 | 0.17 |
ENSDART00000105636
|
myoz1b
|
myozenin 1b |
| chr5_-_12706441 | 0.17 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr13_+_36832145 | 0.17 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr10_+_18953671 | 0.17 |
ENSDART00000135786
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr5_+_50580190 | 0.17 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr17_-_37447917 | 0.16 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr9_-_9000604 | 0.16 |
ENSDART00000145266
|
ing1
|
inhibitor of growth family, member 1 |
| chr14_-_34172522 | 0.16 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr22_-_800449 | 0.16 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr19_+_4835526 | 0.16 |
|
|
|
| chr6_-_3837294 | 0.16 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr23_-_34967879 | 0.16 |
ENSDART00000168731
|
ENSDARG00000061272
|
ENSDARG00000061272 |
| chr17_+_11921705 | 0.16 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr5_-_47523636 | 0.16 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr20_+_27565701 | 0.16 |
|
|
|
| chr8_+_28891430 | 0.16 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr9_-_32348003 | 0.16 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr4_+_4501129 | 0.15 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr20_-_35567985 | 0.15 |
ENSDART00000016090
|
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr5_+_38903966 | 0.15 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr9_-_32347940 | 0.15 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr20_-_3074186 | 0.15 |
ENSDART00000046641
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr9_-_6402663 | 0.15 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr20_-_31594202 | 0.15 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr10_+_18953548 | 0.15 |
ENSDART00000030205
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr20_+_40247918 | 0.15 |
ENSDART00000121818
|
trdn
|
triadin |
| chr12_+_48497255 | 0.15 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr12_+_44829938 | 0.14 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr10_+_20222743 | 0.14 |
ENSDART00000167008
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr9_+_38672040 | 0.14 |
ENSDART00000157556
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr24_-_20496308 | 0.14 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr16_+_53438411 | 0.14 |
ENSDART00000155940
|
CR848746.1
|
ENSDARG00000097164 |
| chr21_-_3462128 | 0.14 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr24_-_24859334 | 0.14 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr18_+_6747156 | 0.14 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr17_-_6294409 | 0.14 |
ENSDART00000064700
|
fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr5_-_64983964 | 0.14 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr10_+_40293111 | 0.14 |
|
|
|
| chr7_+_5855024 | 0.14 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr6_-_10728582 | 0.14 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr25_-_13394261 | 0.14 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr20_+_29788065 | 0.13 |
ENSDART00000164121
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr20_+_4000402 | 0.13 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr6_+_7165326 | 0.13 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr22_-_20899539 | 0.13 |
ENSDART00000100642
|
ell
|
elongation factor RNA polymerase II |
| chr9_-_14172110 | 0.13 |
ENSDART00000108950
|
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr3_-_14981512 | 0.13 |
ENSDART00000131720
|
xpo6
|
exportin 6 |
| chr19_+_5399535 | 0.13 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr14_-_34172619 | 0.13 |
ENSDART00000147380
|
ENSDARG00000092889
|
ENSDARG00000092889 |
| chr20_-_7186902 | 0.13 |
ENSDART00000012247
|
dhcr24
|
24-dehydrocholesterol reductase |
| chr19_-_26188624 | 0.13 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr10_-_36849540 | 0.13 |
|
|
|
| chr25_-_20568599 | 0.13 |
ENSDART00000098076
|
csk
|
c-src tyrosine kinase |
| chr19_-_33356861 | 0.13 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr17_-_5918853 | 0.13 |
ENSDART00000058890
ENSDART00000171084 |
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr1_-_53123905 | 0.13 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr14_+_49875056 | 0.12 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr13_-_36495730 | 0.12 |
ENSDART00000165629
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr23_+_22857958 | 0.12 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr18_+_6664159 | 0.12 |
|
|
|
| chr4_+_12293403 | 0.12 |
ENSDART00000061070
|
mkrn1
|
makorin, ring finger protein, 1 |
| chr22_+_28496424 | 0.12 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr5_-_11471707 | 0.12 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr8_-_17031599 | 0.12 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr17_-_7194180 | 0.12 |
ENSDART00000098731
|
stxbp5b
|
syntaxin binding protein 5b (tomosyn) |
| chr16_+_17762256 | 0.12 |
ENSDART00000128672
|
tmem238b
|
transmembrane protein 238b |
| chr23_-_18131038 | 0.12 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr7_+_55332040 | 0.12 |
ENSDART00000171736
|
trappc2l
|
trafficking protein particle complex 2-like |
| chr25_+_6059393 | 0.12 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr3_-_39163141 | 0.12 |
ENSDART00000102674
ENSDART00000167070 |
plcd3a
|
phospholipase C, delta 3a |
| chr14_-_2288186 | 0.12 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr8_-_1833095 | 0.12 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr13_-_2083873 | 0.12 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr12_-_3630622 | 0.12 |
ENSDART00000164707
|
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr18_-_35861482 | 0.12 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr21_+_36938203 | 0.11 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr1_+_53294559 | 0.11 |
ENSDART00000159900
|
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr25_+_34410718 | 0.11 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr12_-_48092534 | 0.11 |
|
|
|
| chr2_-_14757989 | 0.11 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr7_-_52678749 | 0.11 |
ENSDART00000174329
|
tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr13_-_46423391 | 0.11 |
|
|
|
| chr6_+_4091251 | 0.11 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr5_-_12706568 | 0.11 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr12_-_34786663 | 0.11 |
ENSDART00000158848
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr3_+_44328013 | 0.11 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr25_-_10992022 | 0.11 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr9_-_33116897 | 0.11 |
|
|
|
| chr19_-_1002877 | 0.11 |
|
|
|
| chr16_+_53438473 | 0.11 |
ENSDART00000155940
|
CR848746.1
|
ENSDARG00000097164 |
| chr5_-_23741334 | 0.11 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr5_+_59394192 | 0.10 |
|
|
|
| chr17_+_21944679 | 0.10 |
ENSDART00000063704
|
crip3
|
cysteine-rich protein 3 |
| chr18_+_31020260 | 0.10 |
ENSDART00000154993
|
cd151l
|
CD151 antigen, like |
| chr5_+_26804676 | 0.10 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.3 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.3 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.2 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0050832 | neutrophil mediated immunity(GO:0002446) defense response to fungus(GO:0050832) |
| 0.1 | 0.2 | GO:0086010 | mechanosensory behavior(GO:0007638) membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.2 | GO:0046887 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.2 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.2 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.0 | 0.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0071543 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0098754 | detoxification(GO:0098754) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0072523 | purine-containing compound catabolic process(GO:0072523) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.2 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.1 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.2 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |