Project
DANIO-CODE
Navigation
Downloads
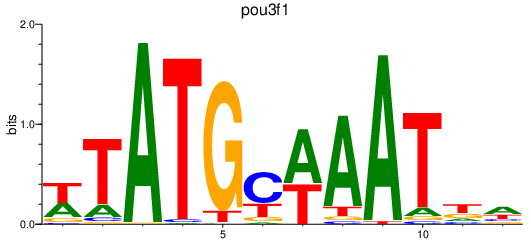
Results for pou3f1
Z-value: 0.82
Transcription factors associated with pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f1
|
ENSDARG00000009823 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f1 | dr10_dc_chr16_+_33639132_33639192 | 0.37 | 1.6e-01 | Click! |
Activity profile of pou3f1 motif
Sorted Z-values of pou3f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_31446156 | 2.23 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr6_-_39315024 | 2.13 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr15_-_16076874 | 1.88 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr3_-_6078015 | 1.80 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr9_-_54126121 | 1.59 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr9_+_54612545 | 1.38 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr14_-_40454194 | 1.35 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr5_+_26829686 | 1.30 |
ENSDART00000087894
|
zgc:153409
|
zgc:153409 |
| chr12_+_23302859 | 1.20 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr13_-_33696425 | 1.20 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr24_-_8590572 | 1.12 |
ENSDART00000082349
|
tfap2a
|
transcription factor AP-2 alpha |
| chr24_-_21005580 | 1.12 |
ENSDART00000154249
|
gramd1c
|
GRAM domain containing 1c |
| chr3_-_44113070 | 1.04 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr18_+_22617072 | 0.99 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr21_+_5781882 | 0.95 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr4_-_9779763 | 0.92 |
ENSDART00000134280
|
svopl
|
SVOP-like |
| chr19_+_3713027 | 0.87 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| KN150001v1_+_14939 | 0.87 |
|
|
|
| chr10_+_2772113 | 0.80 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr15_-_18425091 | 0.80 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr9_-_53898524 | 0.80 |
ENSDART00000175619
ENSDART00000179009 |
CABZ01078244.2
|
ENSDARG00000107473 |
| chr8_+_28584427 | 0.79 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr1_+_53353690 | 0.78 |
ENSDART00000126339
|
dla
|
deltaA |
| chr1_-_24227846 | 0.72 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr5_+_37053530 | 0.72 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr1_-_20131454 | 0.71 |
|
|
|
| chr8_+_13465718 | 0.71 |
ENSDART00000034740
|
fut9d
|
fucosyltransferase 9d |
| chr9_-_43178744 | 0.71 |
|
|
|
| chr7_+_25649559 | 0.71 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr2_+_53253623 | 0.69 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr4_-_28346021 | 0.69 |
ENSDART00000178149
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr22_-_95158 | 0.67 |
|
|
|
| chr13_-_5953330 | 0.64 |
ENSDART00000099224
|
dld
|
deltaD |
| chr2_-_24661477 | 0.64 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr4_+_9177011 | 0.62 |
ENSDART00000057254
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr23_-_6831711 | 0.62 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr17_+_20480419 | 0.60 |
ENSDART00000088490
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr24_-_21005529 | 0.59 |
ENSDART00000154249
|
gramd1c
|
GRAM domain containing 1c |
| chr23_-_3466041 | 0.58 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr18_+_29424354 | 0.57 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr25_-_34619603 | 0.56 |
ENSDART00000114767
|
FQ312024.1
|
ENSDARG00000076129 |
| chr10_+_36096599 | 0.55 |
|
|
|
| chr25_+_34543860 | 0.54 |
ENSDART00000050237
|
hist1h2ba
|
histone cluster 1, H2ba |
| chr13_+_28574593 | 0.54 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr21_+_38770125 | 0.52 |
ENSDART00000149085
|
hnf1bb
|
HNF1 homeobox Bb |
| chr11_+_743048 | 0.51 |
ENSDART00000172972
|
vgll4b
|
vestigial-like family member 4b |
| chr24_-_16995194 | 0.50 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr10_+_32160464 | 0.50 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr7_+_73604595 | 0.49 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr22_+_24617667 | 0.49 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr1_-_16894589 | 0.48 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr2_+_58654570 | 0.48 |
ENSDART00000164102
ENSDART00000158777 |
cirbpa
|
cold inducible RNA binding protein a |
| chr18_+_44656323 | 0.48 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr23_-_26798851 | 0.47 |
ENSDART00000170576
ENSDART00000166123 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr16_-_6298105 | 0.47 |
|
|
|
| chr25_-_34619744 | 0.46 |
ENSDART00000114767
|
FQ312024.1
|
ENSDARG00000076129 |
| chr16_+_53178713 | 0.46 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr7_-_6215188 | 0.46 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr18_+_38768524 | 0.45 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr23_+_28656263 | 0.45 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr1_-_20131665 | 0.43 |
|
|
|
| chr7_+_20699191 | 0.43 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr7_-_19390325 | 0.41 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr6_-_44282665 | 0.41 |
ENSDART00000157215
|
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr1_+_45503061 | 0.40 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr5_-_6004819 | 0.40 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr25_+_19568627 | 0.40 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr4_+_12980657 | 0.39 |
|
|
|
| chr7_+_5839009 | 0.39 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr5_-_53850671 | 0.39 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr23_+_26806694 | 0.39 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr23_+_22858361 | 0.39 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| KN150001v1_+_15017 | 0.39 |
|
|
|
| chr2_-_45375072 | 0.38 |
|
|
|
| chr8_-_12365342 | 0.38 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr11_+_5745644 | 0.38 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr25_-_34607624 | 0.38 |
ENSDART00000128017
|
CU302436.1
|
ENSDARG00000051726 |
| chr25_-_34607515 | 0.37 |
ENSDART00000128017
|
CU302436.1
|
ENSDARG00000051726 |
| chr23_-_4175790 | 0.37 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr6_+_30441419 | 0.37 |
|
|
|
| chr22_-_7371498 | 0.36 |
|
|
|
| chr11_-_35313757 | 0.36 |
ENSDART00000031441
|
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr19_-_30975279 | 0.36 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr24_-_21005426 | 0.35 |
ENSDART00000154249
|
gramd1c
|
GRAM domain containing 1c |
| chr19_-_18689571 | 0.34 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr24_-_16994956 | 0.34 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr1_-_20131393 | 0.34 |
|
|
|
| chr1_+_30992553 | 0.34 |
ENSDART00000112333
|
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr16_+_31890795 | 0.34 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr7_+_5839103 | 0.34 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr7_-_73590802 | 0.33 |
ENSDART00000167855
|
FP236812.8
|
Histone H2B 1/2 |
| chr3_+_23567458 | 0.33 |
ENSDART00000078466
|
hoxb3a
|
homeobox B3a |
| chr14_-_27703812 | 0.32 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr19_-_2486568 | 0.31 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr25_+_34562216 | 0.31 |
ENSDART00000154655
|
CU302436.4
|
ENSDARG00000092743 |
| chr7_-_23601966 | 0.31 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr23_+_11412329 | 0.31 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr12_-_36138709 | 0.30 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr17_+_33766838 | 0.30 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr5_+_4906729 | 0.29 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr25_-_34540270 | 0.29 |
ENSDART00000156508
|
zgc:114046
|
zgc:114046 |
| chr2_-_59019569 | 0.29 |
|
|
|
| chr3_-_6633512 | 0.28 |
ENSDART00000165273
|
GGA3 (1 of many)
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr5_-_41672394 | 0.28 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_-_26799063 | 0.28 |
ENSDART00000157844
|
hspg2
|
heparan sulfate proteoglycan 2 |
| chr23_-_1008307 | 0.28 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr5_-_24806610 | 0.28 |
ENSDART00000089748
|
rorb
|
RAR-related orphan receptor B |
| chr1_-_24227800 | 0.27 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr10_+_36861503 | 0.27 |
ENSDART00000153914
|
BX323076.1
|
ENSDARG00000097240 |
| chr21_-_37404394 | 0.27 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr20_+_27194162 | 0.27 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr12_+_29140448 | 0.26 |
|
|
|
| chr18_+_22228682 | 0.26 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr25_-_14948922 | 0.26 |
ENSDART00000161165
|
pax6a
|
paired box 6a |
| chr3_+_17210396 | 0.26 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr16_+_6694341 | 0.25 |
|
|
|
| chr14_+_6684429 | 0.25 |
ENSDART00000157635
|
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr24_-_18341006 | 0.25 |
ENSDART00000172533
|
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr19_+_20202737 | 0.24 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr13_+_49154065 | 0.24 |
|
|
|
| chr3_-_36126124 | 0.23 |
|
|
|
| chr6_-_42143595 | 0.22 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr19_-_32454747 | 0.22 |
ENSDART00000088358
|
znf704
|
zinc finger protein 704 |
| chr14_-_26406720 | 0.22 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr7_+_10460159 | 0.22 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_+_2772144 | 0.21 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr12_+_46944395 | 0.21 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr16_+_31890895 | 0.21 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr11_-_9479689 | 0.21 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr6_-_33790498 | 0.21 |
|
|
|
| chr13_+_42280697 | 0.21 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_37844454 | 0.20 |
ENSDART00000121768
|
RNaseP_nuc
|
Nuclear RNase P |
| chr5_-_53850436 | 0.20 |
ENSDART00000165346
|
alad
|
aminolevulinate dehydratase |
| chr21_-_26078089 | 0.20 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr16_+_43249142 | 0.19 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr5_+_8642839 | 0.19 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr22_+_4722336 | 0.19 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr16_+_25158949 | 0.19 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr20_-_25727347 | 0.18 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr11_-_9479592 | 0.18 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr1_+_25717717 | 0.18 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr2_-_44402486 | 0.18 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr8_-_12365285 | 0.18 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr1_-_55072375 | 0.17 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr2_-_2109125 | 0.17 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr2_+_58655334 | 0.17 |
|
|
|
| chr23_+_28804842 | 0.17 |
ENSDART00000047378
|
sst3
|
somatostatin 3 |
| chr17_-_44446275 | 0.17 |
ENSDART00000165252
|
slc35f4
|
solute carrier family 35, member F4 |
| chr4_-_9591451 | 0.16 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr14_+_28180174 | 0.16 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr10_-_302737 | 0.16 |
ENSDART00000157582
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr18_+_44656426 | 0.16 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr2_+_55859099 | 0.16 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr15_-_47696481 | 0.16 |
|
|
|
| chr23_+_25224499 | 0.15 |
|
|
|
| chr24_+_16992420 | 0.15 |
|
|
|
| chr15_-_16076807 | 0.15 |
ENSDART00000160114
|
hnf1ba
|
HNF1 homeobox Ba |
| chr21_-_32747483 | 0.15 |
|
|
|
| chr6_+_50393779 | 0.15 |
ENSDART00000055502
|
ergic3
|
ERGIC and golgi 3 |
| chr20_+_48950833 | 0.15 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr25_-_14949028 | 0.15 |
ENSDART00000172538
|
pax6a
|
paired box 6a |
| chr5_-_40961381 | 0.14 |
|
|
|
| chr22_-_38530581 | 0.14 |
ENSDART00000151785
|
klc4
|
kinesin light chain 4 |
| chr21_+_22702293 | 0.14 |
ENSDART00000151342
|
arhgap42a
|
Rho GTPase activating protein 42a |
| chr19_-_43520340 | 0.14 |
|
|
|
| chr16_+_11295005 | 0.14 |
|
|
|
| chr5_+_22632707 | 0.14 |
ENSDART00000003428
|
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr23_+_32085181 | 0.14 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr6_-_54103765 | 0.14 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr5_-_47523737 | 0.14 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr7_-_6330257 | 0.14 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr1_-_1254253 | 0.14 |
ENSDART00000130697
|
runx1
|
runt-related transcription factor 1 |
| chr10_+_3427790 | 0.13 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr16_-_12362566 | 0.13 |
ENSDART00000116542
|
U7
|
U7 small nuclear RNA |
| chr20_+_25727285 | 0.13 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr11_+_18450008 | 0.12 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr7_-_6323808 | 0.11 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr9_-_25151812 | 0.11 |
|
|
|
| chr24_-_37596370 | 0.11 |
ENSDART00000162538
|
cluap1
|
clusterin associated protein 1 |
| chr9_-_1983772 | 0.11 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr7_-_6323664 | 0.11 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr10_-_5820321 | 0.11 |
ENSDART00000172409
|
il6st
|
interleukin 6 signal transducer |
| chr7_+_58427742 | 0.10 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr3_+_33213672 | 0.10 |
ENSDART00000162870
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr3_-_32205873 | 0.10 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr10_+_10428297 | 0.10 |
ENSDART00000179214
|
sardh
|
sarcosine dehydrogenase |
| chr21_+_19311031 | 0.09 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr17_+_7438152 | 0.09 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr12_+_16038265 | 0.08 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr23_-_20332076 | 0.08 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr10_-_26263737 | 0.08 |
ENSDART00000079194
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr3_+_1338347 | 0.07 |
ENSDART00000092690
ENSDART00000165395 |
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr22_+_18218979 | 0.07 |
|
|
|
| chr12_+_48497255 | 0.07 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr23_+_21453261 | 0.07 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr20_+_35954442 | 0.07 |
ENSDART00000102611
|
cd2ap
|
CD2-associated protein |
| chr23_-_36812319 | 0.07 |
ENSDART00000109976
|
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr7_-_24428781 | 0.06 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr13_-_49153890 | 0.06 |
ENSDART00000136991
|
irf2bp2a
|
interferon regulatory factor 2 binding protein 2a |
| chr14_+_16460336 | 0.06 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.4 | 1.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.2 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.3 | 1.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.0 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 0.7 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.2 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 0.8 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 0.6 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.1 | 1.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.6 | GO:0045639 | positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.1 | 1.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.5 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.1 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.1 | 0.2 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.1 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.4 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |