Project
DANIO-CODE
Navigation
Downloads
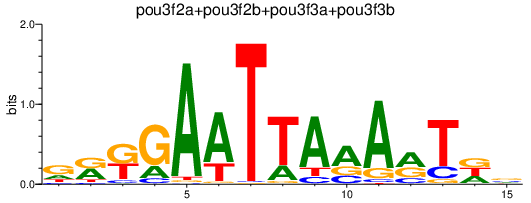
Results for pou3f2a+pou3f2b+pou3f3a+pou3f3b
Z-value: 1.00
Transcription factors associated with pou3f2a+pou3f2b+pou3f3a+pou3f3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f3a
|
ENSDARG00000042032 | POU class 3 homeobox 3a |
|
pou3f2a
|
ENSDARG00000070220 | POU class 3 homeobox 2a |
|
pou3f2b
|
ENSDARG00000076262 | POU class 3 homeobox 2b |
|
pou3f3b
|
ENSDARG00000095896 | POU class 3 homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f2a | dr10_dc_chr4_+_5733160_5733232 | -0.84 | 4.9e-05 | Click! |
| pou3f2b | dr10_dc_chr16_+_32605735_32605856 | -0.83 | 7.4e-05 | Click! |
| pou3f3b | dr10_dc_chr6_+_14823185_14823275 | -0.82 | 1.0e-04 | Click! |
| pou3f3a | dr10_dc_chr9_-_6683651_6683699 | -0.73 | 1.4e-03 | Click! |
Activity profile of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
Sorted Z-values of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f2a+pou3f2b+pou3f3a+pou3f3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_34150130 | 2.30 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr17_+_10582214 | 2.07 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr2_-_2373601 | 1.77 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr13_+_35799681 | 1.70 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_+_35799602 | 1.62 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr20_+_34091702 | 1.58 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr20_-_9135267 | 1.57 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr1_-_54391298 | 1.55 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr10_-_7513764 | 1.53 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr24_+_34183462 | 1.51 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr1_-_8593850 | 1.49 |
ENSDART00000146065
ENSDART00000114876 |
ubn1
|
ubinuclein 1 |
| chr14_-_33641145 | 1.47 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr7_-_8466131 | 1.46 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr1_+_13244145 | 1.43 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr24_-_36413059 | 1.40 |
ENSDART00000062736
|
coasy
|
CoA synthase |
| chr7_-_5253629 | 1.39 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr8_-_49227232 | 1.36 |
|
|
|
| chr25_+_5845303 | 1.35 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr5_+_26888744 | 1.32 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr6_+_28218420 | 1.27 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr3_+_32617947 | 1.27 |
ENSDART00000125260
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr8_-_49227286 | 1.20 |
|
|
|
| chr24_+_10758290 | 1.18 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr3_+_32285237 | 1.17 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr24_+_34183557 | 1.16 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr11_+_39664664 | 1.16 |
ENSDART00000102734
ENSDART00000137516 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr1_-_39141284 | 1.16 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr15_+_14656645 | 1.15 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr17_+_24790812 | 1.14 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr22_+_15317622 | 1.13 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr24_-_25297360 | 1.12 |
ENSDART00000138215
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_13244109 | 1.12 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr21_+_22598805 | 1.12 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr16_-_33047066 | 1.12 |
ENSDART00000147941
ENSDART00000075218 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr3_+_40654510 | 1.11 |
ENSDART00000014729
|
arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr24_-_36412892 | 1.10 |
ENSDART00000148868
|
coasy
|
CoA synthase |
| chr17_+_24791024 | 1.09 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr9_-_7308912 | 1.08 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr23_+_46000362 | 1.07 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr16_-_47446494 | 1.06 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr19_+_20617380 | 1.04 |
ENSDART00000133633
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr2_-_47766563 | 1.04 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr2_-_3588636 | 1.04 |
ENSDART00000055618
|
iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr18_+_8959686 | 1.03 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr7_+_38444768 | 1.02 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr18_+_27457678 | 1.01 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr5_-_39205454 | 0.98 |
ENSDART00000133231
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr15_-_5913264 | 0.97 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr11_+_34909393 | 0.96 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr15_+_14656797 | 0.96 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr18_+_6747156 | 0.95 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr2_+_15379717 | 0.95 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr3_-_20912934 | 0.95 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr21_+_19040595 | 0.95 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr9_+_23813071 | 0.95 |
|
|
|
| chr10_-_21404605 | 0.95 |
ENSDART00000125167
|
avd
|
avidin |
| chr18_+_15137751 | 0.94 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr14_+_9175399 | 0.94 |
ENSDART00000054690
ENSDART00000135449 |
st3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr19_+_33551956 | 0.94 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr2_-_26941084 | 0.93 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_-_26941232 | 0.93 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr21_-_30247820 | 0.92 |
ENSDART00000066363
|
ENSDARG00000045127
|
ENSDARG00000045127 |
| chr13_+_11696495 | 0.92 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr17_+_4902984 | 0.92 |
ENSDART00000064313
ENSDART00000121806 |
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr20_+_51193122 | 0.91 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr24_-_6304289 | 0.91 |
ENSDART00000140212
|
zgc:174877
|
zgc:174877 |
| chr16_+_53632289 | 0.91 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr3_-_33296077 | 0.91 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr13_-_17991273 | 0.91 |
ENSDART00000128748
|
fam21c
|
family with sequence similarity 21, member C |
| chr15_+_20054306 | 0.90 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr4_+_20074260 | 0.90 |
ENSDART00000024925
|
gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr14_+_25208174 | 0.90 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr25_+_7067115 | 0.89 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr5_-_37785264 | 0.87 |
|
|
|
| chr24_+_21030141 | 0.87 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr25_-_3619590 | 0.86 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr7_-_8465988 | 0.85 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr25_+_33118422 | 0.85 |
|
|
|
| chr23_+_44470297 | 0.85 |
ENSDART00000149842
|
MEPCE
|
methylphosphate capping enzyme |
| chr13_-_49749441 | 0.84 |
ENSDART00000136165
|
lyst
|
lysosomal trafficking regulator |
| chr21_+_6008781 | 0.84 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr1_-_8843231 | 0.84 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr19_+_20617246 | 0.83 |
ENSDART00000133633
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr22_+_8856370 | 0.83 |
ENSDART00000178799
|
CABZ01046433.1
|
ENSDARG00000109049 |
| chr2_-_6153363 | 0.83 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr12_+_31345948 | 0.82 |
ENSDART00000031154
|
gucy2g
|
guanylate cyclase 2g |
| chr1_-_22617455 | 0.82 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr23_-_3568408 | 0.82 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr19_+_7489293 | 0.82 |
ENSDART00000081746
|
apoa1bp
|
apolipoprotein A-I binding protein |
| chr2_+_30497550 | 0.81 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr1_+_48770997 | 0.81 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr24_+_36451347 | 0.81 |
ENSDART00000142264
|
grnb
|
granulin b |
| chr8_-_22493608 | 0.81 |
ENSDART00000021514
|
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr14_+_29932533 | 0.81 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr17_-_49980590 | 0.80 |
ENSDART00000106562
|
zgc:113886
|
zgc:113886 |
| chr5_+_29193876 | 0.80 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr8_-_28323975 | 0.79 |
|
|
|
| chr3_+_54551887 | 0.79 |
ENSDART00000169663
|
CT573860.1
|
ENSDARG00000098327 |
| chr20_+_31036284 | 0.79 |
ENSDART00000153344
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr3_+_37648476 | 0.79 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr3_+_32617855 | 0.79 |
ENSDART00000053684
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr25_+_2252667 | 0.78 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr9_-_21287478 | 0.77 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr9_-_34700736 | 0.77 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr2_-_37299150 | 0.76 |
ENSDART00000137598
|
nadkb
|
NAD kinase b |
| chr10_+_10429005 | 0.76 |
ENSDART00000129253
ENSDART00000131043 |
sardh
|
sarcosine dehydrogenase |
| chr25_-_36512943 | 0.76 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr17_+_13803088 | 0.76 |
ENSDART00000176632
|
BX005443.1
|
ENSDARG00000107833 |
| chr2_-_19705537 | 0.76 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr1_-_8842947 | 0.75 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr17_-_27217309 | 0.75 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr23_-_17503357 | 0.75 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr18_-_7138171 | 0.74 |
ENSDART00000149122
|
cfap161
|
cilia and flagella associated protein 161 |
| chr3_+_17783319 | 0.74 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr13_+_35799851 | 0.74 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_-_50940551 | 0.74 |
|
|
|
| chr17_-_18878230 | 0.73 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr23_-_41367783 | 0.73 |
|
|
|
| chr5_-_8177663 | 0.73 |
ENSDART00000168659
|
zgc:153352
|
zgc:153352 |
| chr17_-_23234771 | 0.73 |
|
|
|
| chr13_+_29795813 | 0.73 |
ENSDART00000131609
|
cuedc2
|
CUE domain containing 2 |
| chr2_+_15379961 | 0.72 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr18_+_15137711 | 0.72 |
ENSDART00000128609
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr8_+_2484943 | 0.72 |
ENSDART00000101137
|
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr14_-_16469435 | 0.72 |
ENSDART00000113711
|
zgc:123335
|
zgc:123335 |
| chr25_-_14684924 | 0.72 |
|
|
|
| chr20_-_23354440 | 0.71 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
| chr16_-_52090636 | 0.71 |
ENSDART00000155308
|
FO704712.1
|
ENSDARG00000063300 |
| chr21_-_31215463 | 0.71 |
ENSDART00000170507
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr23_-_27768575 | 0.71 |
ENSDART00000146703
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr2_-_37116119 | 0.71 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr13_+_51747107 | 0.70 |
ENSDART00000161098
|
lclat1
|
lysocardiolipin acyltransferase 1 |
| chr5_+_36049836 | 0.70 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr12_-_26529300 | 0.69 |
ENSDART00000162163
ENSDART00000087067 |
hexdc
|
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr4_-_24033285 | 0.69 |
ENSDART00000132855
|
cpm
|
carboxypeptidase M |
| chr15_-_25333252 | 0.69 |
ENSDART00000127771
|
mettl16
|
methyltransferase like 16 |
| chr10_+_44456550 | 0.69 |
ENSDART00000157611
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr4_-_20119812 | 0.69 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr24_+_29902165 | 0.69 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr7_-_38443332 | 0.68 |
ENSDART00000047220
|
fbxo3
|
F-box protein 3 |
| chr2_-_26940965 | 0.68 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr7_-_15008685 | 0.68 |
ENSDART00000173048
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr14_+_9175488 | 0.67 |
ENSDART00000054690
|
st3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr2_-_44123230 | 0.67 |
|
|
|
| chr19_+_27338879 | 0.67 |
ENSDART00000149988
|
nelfe
|
negative elongation factor complex member E |
| chr11_+_30006715 | 0.67 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr24_-_23570846 | 0.67 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr21_-_30247906 | 0.67 |
ENSDART00000066363
|
ENSDARG00000045127
|
ENSDARG00000045127 |
| chr5_-_57053687 | 0.66 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr7_-_51474798 | 0.66 |
ENSDART00000175523
|
hdac8
|
histone deacetylase 8 |
| chr15_-_14102102 | 0.66 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr21_+_15637516 | 0.66 |
ENSDART00000149371
|
idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr25_+_30627892 | 0.66 |
|
|
|
| chr10_-_25366450 | 0.65 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr12_-_19063761 | 0.65 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr12_+_19837053 | 0.65 |
ENSDART00000015780
|
ercc4
|
excision repair cross-complementation group 4 |
| chr21_+_26954898 | 0.65 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr22_-_26269585 | 0.65 |
ENSDART00000043774
|
sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr4_-_13932592 | 0.65 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr8_-_13231600 | 0.64 |
ENSDART00000014528
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr3_-_35735315 | 0.64 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr13_-_50940520 | 0.64 |
|
|
|
| chr7_-_51474531 | 0.64 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr5_+_23582676 | 0.64 |
ENSDART00000139520
|
tp53
|
tumor protein p53 |
| chr17_+_21797726 | 0.63 |
ENSDART00000079011
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr16_-_7703633 | 0.63 |
ENSDART00000148581
|
CT574585.1
|
ENSDARG00000095852 |
| chr7_-_25991514 | 0.63 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr12_+_4651222 | 0.63 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr5_-_68408107 | 0.63 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr7_-_24604255 | 0.63 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr14_+_30390987 | 0.63 |
|
|
|
| chr13_-_36635434 | 0.63 |
ENSDART00000179302
|
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_-_49256667 | 0.63 |
ENSDART00000112806
|
orc3
|
origin recognition complex, subunit 3 |
| chr7_+_22042296 | 0.63 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr8_-_21110183 | 0.62 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr13_-_49886891 | 0.62 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr23_-_29952057 | 0.62 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr23_-_32173435 | 0.62 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr15_-_33342455 | 0.62 |
|
|
|
| chr1_-_46423056 | 0.61 |
ENSDART00000053157
|
setd4
|
SET domain containing 4 |
| chr1_+_39141680 | 0.61 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr23_-_17503329 | 0.61 |
ENSDART00000132024
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr7_-_8465929 | 0.61 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr20_+_37592206 | 0.61 |
ENSDART00000153092
|
BX088686.1
|
ENSDARG00000096774 |
| chr7_+_21485734 | 0.60 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr3_+_16692138 | 0.60 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr2_+_44041230 | 0.60 |
ENSDART00000143885
|
gbp3
|
guanylate binding protein 3 |
| chr6_+_11162062 | 0.60 |
ENSDART00000151548
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr20_+_32620786 | 0.60 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr13_+_29795897 | 0.60 |
ENSDART00000131609
|
cuedc2
|
CUE domain containing 2 |
| chr10_+_20435169 | 0.60 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr17_-_31802796 | 0.60 |
ENSDART00000172519
|
abraxas2b
|
abraxas 2b, BRISC complex subunit |
| chr7_+_5789700 | 0.59 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr19_-_26188747 | 0.59 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr21_-_7322856 | 0.59 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr19_-_30834980 | 0.59 |
ENSDART00000043554
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr23_-_24616306 | 0.58 |
ENSDART00000088777
|
atp13a2
|
ATPase type 13A2 |
| chr4_+_72009475 | 0.58 |
ENSDART00000172536
|
znf989
|
zinc finger protein 989 |
| chr13_-_6195363 | 0.58 |
ENSDART00000148907
|
mcph1
|
microcephalin 1 |
| chr3_-_141976 | 0.58 |
ENSDART00000114289
|
ENSDARG00000075737
|
ENSDARG00000075737 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0051977 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.7 | 2.8 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.5 | 1.5 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.4 | 1.2 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.4 | 1.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 1.0 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.3 | 1.2 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.3 | 1.7 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.3 | 1.0 | GO:0046100 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.2 | 0.7 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.2 | 1.0 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 1.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 0.9 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.2 | 1.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 0.9 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.2 | 0.8 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.2 | 1.0 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.8 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 0.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 0.8 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.2 | 2.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.5 | GO:0097704 | response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.2 | 1.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.6 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.5 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0060217 | phosphatidylinositol acyl-chain remodeling(GO:0036149) hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.1 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.6 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 1.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.9 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.1 | 0.7 | GO:0060296 | regulation of cilium beat frequency(GO:0003356) regulation of translational termination(GO:0006449) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.4 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.9 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.6 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.6 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.5 | GO:1903449 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 0.1 | 0.4 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.6 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.7 | GO:0098869 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.3 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.1 | 1.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.4 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.5 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.7 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.1 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.3 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.1 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 3.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0071674 | positive regulation of lamellipodium assembly(GO:0010592) regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) negative regulation of skeletal muscle cell proliferation(GO:0014859) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.8 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0060232 | delamination(GO:0060232) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.0 | 0.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.6 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.8 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.5 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.2 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 1.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 2.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.1 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.7 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.3 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.3 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 0.6 | GO:1990077 | primosome complex(GO:1990077) |
| 0.2 | 1.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.5 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 1.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.5 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.2 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 1.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.4 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 1.0 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 1.0 | 2.9 | GO:0097020 | COPII adaptor activity(GO:0097020) |
| 0.8 | 2.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 1.6 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.4 | 1.6 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.4 | 1.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.0 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.3 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 0.8 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.3 | 1.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 0.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.7 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.2 | 1.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 0.7 | GO:0052907 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.2 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.8 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.2 | 1.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 2.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.5 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 1.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 0.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.6 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 2.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.3 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 0.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.6 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.9 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.0 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |