Project
DANIO-CODE
Navigation
Downloads
Results for pou4f2
Z-value: 0.56
Transcription factors associated with pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f2
|
ENSDARG00000069737 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f2 | dr10_dc_chr1_+_35704610_35704630 | 0.65 | 6.5e-03 | Click! |
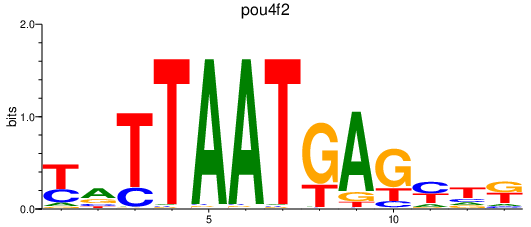
Activity profile of pou4f2 motif
Sorted Z-values of pou4f2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15425559 | 1.17 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_-_30015572 | 0.98 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr16_+_46145286 | 0.76 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr24_-_21778717 | 0.76 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr8_+_15989815 | 0.76 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr6_+_27156169 | 0.70 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr7_+_32097831 | 0.64 |
ENSDART00000169588
|
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr1_+_46537108 | 0.63 |
|
|
|
| chr22_-_13018196 | 0.63 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr15_-_31687 | 0.61 |
|
|
|
| chr16_-_12282596 | 0.59 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr23_+_22763861 | 0.56 |
|
|
|
| chr23_+_22763789 | 0.55 |
|
|
|
| chr13_+_35213326 | 0.53 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr25_+_30472954 | 0.49 |
|
|
|
| chr7_+_73408688 | 0.49 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr25_+_30473054 | 0.47 |
|
|
|
| chr1_-_50215233 | 0.47 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr3_-_28120668 | 0.45 |
|
|
|
| chr16_+_51319421 | 0.44 |
|
|
|
| chr5_+_17120453 | 0.42 |
|
|
|
| chr3_-_30303173 | 0.40 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr17_-_12772433 | 0.39 |
ENSDART00000044126
|
ENSDARG00000020655
|
ENSDARG00000020655 |
| chr4_-_1346961 | 0.39 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr11_-_42626842 | 0.38 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr5_-_71643185 | 0.37 |
ENSDART00000029014
|
pax8
|
paired box 8 |
| chr5_-_62821458 | 0.37 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr15_-_46670611 | 0.37 |
|
|
|
| chr9_-_32942783 | 0.36 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr15_-_24987099 | 0.34 |
|
|
|
| chr11_+_38013238 | 0.34 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr3_+_43010408 | 0.33 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr6_+_3873114 | 0.32 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr6_-_43094573 | 0.32 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr25_+_6059393 | 0.32 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr8_-_27839244 | 0.32 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr14_+_46896395 | 0.31 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr6_+_39186673 | 0.31 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr8_-_15091823 | 0.31 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr20_-_38714872 | 0.30 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr12_-_10372055 | 0.30 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr3_-_28120760 | 0.30 |
|
|
|
| chr3_+_47735048 | 0.29 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr21_+_37668334 | 0.29 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr1_+_53127046 | 0.29 |
|
|
|
| chr1_-_18581114 | 0.28 |
ENSDART00000178278
|
BX546494.1
|
ENSDARG00000107606 |
| KN150670v1_-_72156 | 0.28 |
|
|
|
| chr23_+_22763739 | 0.28 |
|
|
|
| chr2_-_44329092 | 0.28 |
|
|
|
| chr6_+_54179493 | 0.27 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr20_+_54595334 | 0.27 |
|
|
|
| chr20_-_18836593 | 0.26 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr1_-_37475807 | 0.26 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr1_+_13779755 | 0.25 |
|
|
|
| chr9_-_39190534 | 0.25 |
|
|
|
| chr7_+_73408746 | 0.25 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr21_+_36938230 | 0.23 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr17_+_26946957 | 0.23 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr23_+_25378921 | 0.23 |
ENSDART00000143291
|
RPL41
|
ribosomal protein L41 |
| chr3_-_58739476 | 0.22 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr10_+_39291358 | 0.22 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr11_-_44724371 | 0.22 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr2_+_29265967 | 0.21 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr18_+_1353857 | 0.19 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr12_+_24833569 | 0.18 |
ENSDART00000014868
|
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr17_-_37447917 | 0.17 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr6_-_8009055 | 0.17 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr8_-_4270732 | 0.16 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr16_+_11295005 | 0.15 |
|
|
|
| chr24_+_26287471 | 0.15 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr8_+_6533379 | 0.15 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr6_-_43094926 | 0.13 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr18_+_6664159 | 0.13 |
|
|
|
| chr2_-_26160882 | 0.13 |
ENSDART00000133163
|
CR392026.2
|
ENSDARG00000094803 |
| chr19_+_1743359 | 0.13 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr24_-_41403824 | 0.13 |
ENSDART00000063504
ENSDART00000169374 |
xylb
|
xylulokinase homolog (H. influenzae) |
| chr18_-_40518772 | 0.12 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr13_-_46423391 | 0.12 |
|
|
|
| chr2_-_40170303 | 0.11 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr3_-_58739399 | 0.11 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr13_-_42274486 | 0.10 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr9_+_38278401 | 0.10 |
ENSDART00000123749
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr4_-_2707563 | 0.09 |
ENSDART00000021953
ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr15_-_56306 | 0.09 |
ENSDART00000168340
|
CABZ01080370.1
|
POU class 2 homeobox 1a |
| chr15_+_43192811 | 0.09 |
|
|
|
| chr11_+_44240444 | 0.09 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr17_-_12812747 | 0.09 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr7_-_52848084 | 0.09 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr8_+_8889274 | 0.07 |
ENSDART00000142075
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr23_-_17524325 | 0.06 |
ENSDART00000104680
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr2_+_36957471 | 0.06 |
ENSDART00000143915
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr13_+_36496475 | 0.06 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr7_+_17527276 | 0.06 |
|
|
|
| chr7_-_7530898 | 0.06 |
ENSDART00000113131
|
intu
|
inturned planar cell polarity protein |
| chr22_-_342682 | 0.05 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr7_-_17527827 | 0.05 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr19_-_5186692 | 0.05 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr17_-_20182801 | 0.03 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr4_+_9835529 | 0.03 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr4_+_13430131 | 0.03 |
ENSDART00000137200
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr20_-_18836698 | 0.02 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr2_-_10020770 | 0.02 |
ENSDART00000046587
|
ap2m1a
|
adaptor-related protein complex 2, mu 1 subunit, a |
| chr9_-_50304120 | 0.02 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr18_-_15582896 | 0.01 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr17_-_37447869 | 0.01 |
ENSDART00000148160
|
crip1
|
cysteine-rich protein 1 |
| chr8_+_17132156 | 0.01 |
ENSDART00000134665
|
cenph
|
centromere protein H |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.6 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.3 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.5 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.4 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.3 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.1 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |