Project
DANIO-CODE
Navigation
Downloads
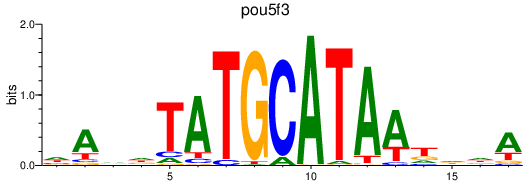
Results for pou5f3
Z-value: 0.33
Transcription factors associated with pou5f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou5f3
|
ENSDARG00000044774 | POU domain, class 5, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou5f3 | dr10_dc_chr21_-_13593659_13594156 | -0.03 | 9.1e-01 | Click! |
Activity profile of pou5f3 motif
Sorted Z-values of pou5f3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou5f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_33783345 | 0.59 |
ENSDART00000143333
|
pou6f1
|
POU class 6 homeobox 1 |
| chr1_-_54391298 | 0.57 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr24_-_20496410 | 0.53 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr22_+_24532560 | 0.46 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr24_-_20496308 | 0.42 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr23_-_33783396 | 0.34 |
ENSDART00000130338
|
pou6f1
|
POU class 6 homeobox 1 |
| chr7_+_24373948 | 0.33 |
ENSDART00000150233
|
gba2
|
glucosidase, beta (bile acid) 2 |
| chr8_+_41004169 | 0.27 |
|
|
|
| chr7_-_24373579 | 0.24 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr17_+_28653205 | 0.23 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr23_+_33981443 | 0.21 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr5_-_47523737 | 0.21 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr5_-_47523636 | 0.18 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr1_+_22583970 | 0.18 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr8_+_41003546 | 0.15 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr23_+_33981358 | 0.14 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr17_+_28653236 | 0.11 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr9_+_12473243 | 0.11 |
ENSDART00000132709
|
tmem41aa
|
transmembrane protein 41aa |
| chr18_-_36291663 | 0.10 |
|
|
|
| chr24_-_24859135 | 0.09 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr22_+_15480482 | 0.08 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr7_+_60745321 | 0.07 |
|
|
|
| chr11_+_18897913 | 0.07 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr16_-_31393328 | 0.06 |
ENSDART00000178298
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr24_-_24859334 | 0.06 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr4_-_5293599 | 0.05 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr16_-_42619967 | 0.04 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr18_+_7584201 | 0.04 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr10_-_44713414 | 0.04 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr11_+_18897769 | 0.03 |
ENSDART00000163006
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr8_-_16556749 | 0.03 |
ENSDART00000101655
|
calr
|
calreticulin |
| KN149781v1_+_4885 | 0.00 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |