Project
DANIO-CODE
Navigation
Downloads
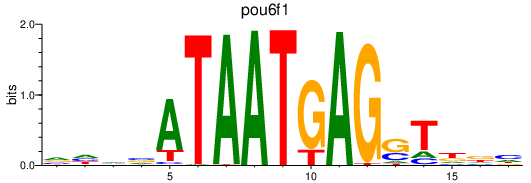
Results for pou6f1
Z-value: 0.58
Transcription factors associated with pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f1
|
ENSDARG00000011570 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f1 | dr10_dc_chr23_-_33783345_33783373 | -0.83 | 5.7e-05 | Click! |
Activity profile of pou6f1 motif
Sorted Z-values of pou6f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_56970554 | 1.88 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr9_-_14533551 | 1.46 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr19_-_31815128 | 1.40 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr23_+_35819625 | 1.35 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr4_-_76620869 | 1.05 |
|
|
|
| KN150207v1_-_1208 | 1.01 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr21_-_22510751 | 1.01 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr22_-_13325409 | 0.95 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr5_-_13872815 | 0.93 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr22_-_336857 | 0.90 |
|
|
|
| chr13_+_21989327 | 0.83 |
ENSDART00000173206
|
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr1_+_13779755 | 0.80 |
|
|
|
| chr22_+_25652275 | 0.78 |
ENSDART00000087769
|
si:ch211-250e5.2
|
si:ch211-250e5.2 |
| chr13_+_371940 | 0.76 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr15_-_17933972 | 0.76 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr12_+_20230575 | 0.74 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr7_-_46510632 | 0.73 |
|
|
|
| chr22_+_16509286 | 0.73 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr9_-_30437160 | 0.71 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr1_+_51706536 | 0.70 |
|
|
|
| chr19_-_35513216 | 0.66 |
|
|
|
| chr7_+_25794014 | 0.64 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr25_+_36878565 | 0.61 |
ENSDART00000157596
|
ric3b
|
RIC3 acetylcholine receptor chaperone b |
| chr22_-_337198 | 0.59 |
|
|
|
| chr19_-_22182031 | 0.57 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr9_-_785511 | 0.57 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr2_-_44402486 | 0.55 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr3_+_21059221 | 0.55 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr8_+_14120313 | 0.54 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr9_-_41982635 | 0.54 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr7_-_72500748 | 0.53 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr17_+_12544451 | 0.51 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr19_+_42899678 | 0.51 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr10_+_2688361 | 0.50 |
ENSDART00000130793
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr15_-_33782909 | 0.49 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr24_-_23155932 | 0.49 |
|
|
|
| chr11_-_6442836 | 0.47 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr16_-_11908084 | 0.47 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr4_+_12616987 | 0.46 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr4_+_12617165 | 0.46 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr13_+_18401965 | 0.46 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr6_+_60060297 | 0.45 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr9_-_22528568 | 0.45 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr22_-_13325475 | 0.45 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr22_-_336942 | 0.43 |
|
|
|
| chr11_-_7310284 | 0.42 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr18_-_50527399 | 0.42 |
ENSDART00000033591
|
cd276
|
CD276 molecule |
| chr12_+_18559530 | 0.41 |
ENSDART00000152948
|
rgs9b
|
regulator of G protein signaling 9b |
| chr18_+_34205362 | 0.41 |
ENSDART00000130831
|
gmps
|
guanine monophosphate synthase |
| chr7_-_9557990 | 0.40 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr1_+_51706568 | 0.40 |
|
|
|
| chr4_-_1564624 | 0.38 |
ENSDART00000168633
|
BICD1 (1 of many)
|
BICD cargo adaptor 1 |
| chr9_-_785569 | 0.38 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr8_+_14120357 | 0.38 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr17_-_43725003 | 0.36 |
|
|
|
| chr7_+_25793777 | 0.35 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr13_+_43266551 | 0.35 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr13_-_11553609 | 0.35 |
|
|
|
| chr5_-_38022706 | 0.34 |
|
|
|
| chr2_+_29265967 | 0.32 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr5_-_70915664 | 0.31 |
|
|
|
| chr7_-_14937467 | 0.29 |
ENSDART00000031049
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr8_+_37136126 | 0.28 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr2_-_44402452 | 0.27 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr14_+_45619218 | 0.27 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr19_+_21781738 | 0.26 |
|
|
|
| chr17_-_49355712 | 0.26 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr9_-_785400 | 0.26 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr22_+_11490729 | 0.25 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr10_-_24422496 | 0.25 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr13_+_21989216 | 0.25 |
ENSDART00000165842
|
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr2_-_50638153 | 0.25 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein-like 2b |
| chr5_-_13872772 | 0.24 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr4_-_13394059 | 0.23 |
ENSDART00000175909
ENSDART00000174861 |
BX511038.1
|
ENSDARG00000108089 |
| chr18_+_1353738 | 0.23 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr4_-_74789596 | 0.23 |
ENSDART00000174356
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr3_-_8608620 | 0.22 |
|
|
|
| chr1_-_24493345 | 0.22 |
ENSDART00000177225
|
tmem154
|
transmembrane protein 154 |
| chr17_+_1986151 | 0.21 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr23_-_36198253 | 0.20 |
|
|
|
| chr2_+_6331176 | 0.20 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr2_-_21323717 | 0.19 |
ENSDART00000113384
|
lyrm4
|
LYR motif containing 4 |
| chr10_-_45212226 | 0.19 |
ENSDART00000170413
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr14_-_28260272 | 0.18 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr18_-_15582896 | 0.18 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr23_+_46015863 | 0.17 |
ENSDART00000012234
|
syap1
|
synapse associated protein 1 |
| chr19_-_11161797 | 0.17 |
|
|
|
| chr4_+_17088146 | 0.16 |
|
|
|
| chr25_+_6059393 | 0.16 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr15_-_5732737 | 0.15 |
ENSDART00000109599
|
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr16_-_5254337 | 0.15 |
ENSDART00000134981
|
dctn3
|
dynactin 3 (p22) |
| chr6_-_43782889 | 0.13 |
|
|
|
| chr12_-_22387786 | 0.11 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr5_+_21593388 | 0.11 |
ENSDART00000090375
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr8_+_30446599 | 0.11 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr22_+_34735496 | 0.09 |
ENSDART00000155906
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_+_46631965 | 0.08 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr17_-_49355906 | 0.08 |
ENSDART00000162563
|
znf292a
|
zinc finger protein 292a |
| chr6_-_9440411 | 0.08 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr3_+_21059313 | 0.06 |
ENSDART00000141410
|
zgc:123295
|
zgc:123295 |
| chr25_+_7287952 | 0.05 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr14_+_47016666 | 0.05 |
ENSDART00000145241
|
slu7
|
SLU7 homolog, splicing factor |
| chr23_-_17524325 | 0.05 |
ENSDART00000104680
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr18_+_1353857 | 0.04 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr9_+_41910191 | 0.04 |
|
|
|
| chr12_+_48176075 | 0.03 |
|
|
|
| chr5_-_39136238 | 0.03 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr21_-_20295921 | 0.02 |
ENSDART00000166049
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 0.8 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.2 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 1.5 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 1.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.5 | GO:0046327 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 1.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |