Project
DANIO-CODE
Navigation
Downloads
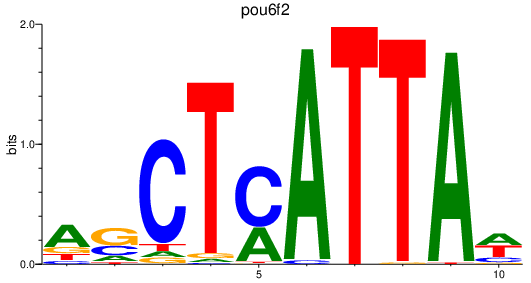
Results for pou6f2
Z-value: 0.93
Transcription factors associated with pou6f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f2
|
ENSDARG00000086362 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f2 | dr10_dc_chr24_+_9898743_9898830 | -0.90 | 2.8e-06 | Click! |
Activity profile of pou6f2 motif
Sorted Z-values of pou6f2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_56970554 | 3.16 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr8_-_26623251 | 3.13 |
ENSDART00000159264
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr23_+_35819625 | 2.96 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr13_+_35213326 | 2.69 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr18_+_5899355 | 2.62 |
|
|
|
| chr12_-_17576409 | 2.30 |
ENSDART00000147282
|
pvalb9
|
parvalbumin 9 |
| chr20_-_22576513 | 2.25 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr19_+_31046291 | 2.11 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr6_+_3873114 | 2.05 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr15_-_31687 | 1.97 |
|
|
|
| chr3_-_6078015 | 1.97 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr24_-_21778717 | 1.94 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr22_-_13325409 | 1.90 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr12_-_17576531 | 1.88 |
ENSDART00000105974
|
pvalb9
|
parvalbumin 9 |
| chr3_-_60929921 | 1.81 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr9_-_14533551 | 1.72 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr3_-_42649955 | 1.68 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr3_-_32202533 | 1.68 |
ENSDART00000155757
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr6_-_43094573 | 1.60 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr15_-_4537178 | 1.55 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr19_-_31815128 | 1.55 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr21_-_39132283 | 1.54 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr7_-_31488479 | 1.53 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr2_+_55888093 | 1.50 |
ENSDART00000146160
|
loxl5b
|
lysyl oxidase-like 5b |
| chr17_+_25503946 | 1.49 |
|
|
|
| chr13_+_24531753 | 1.48 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr9_-_30437160 | 1.45 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr4_-_17640519 | 1.44 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr12_-_26314881 | 1.42 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr2_-_44402486 | 1.39 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr12_+_20230575 | 1.37 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr11_-_27455242 | 1.37 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr4_-_15442828 | 1.34 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr24_+_24308055 | 1.33 |
|
|
|
| chr1_+_13779755 | 1.32 |
|
|
|
| chr6_+_27156169 | 1.25 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr3_-_42649907 | 1.24 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr9_-_32942783 | 1.24 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr25_-_14472107 | 1.23 |
|
|
|
| chr15_-_17933972 | 1.23 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr2_-_35584440 | 1.21 |
ENSDART00000125298
|
tnw
|
tenascin W |
| chr24_+_9603813 | 1.18 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr11_-_37158131 | 1.17 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr16_+_46145286 | 1.17 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr3_-_56474209 | 1.15 |
ENSDART00000156398
|
si:ch211-189a21.1
|
si:ch211-189a21.1 |
| chr11_+_19894772 | 1.15 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr23_+_31181140 | 1.14 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr13_-_22515619 | 1.14 |
ENSDART00000136808
ENSDART00000131262 |
mmrn2a
|
multimerin 2a |
| chr17_-_48623315 | 1.13 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr7_-_46510632 | 1.12 |
|
|
|
| chr3_-_60930025 | 1.10 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr5_-_13872815 | 1.10 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr19_+_48499602 | 1.09 |
|
|
|
| chr23_-_29138952 | 1.09 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr20_+_26408687 | 1.07 |
|
|
|
| chr6_+_16279737 | 1.05 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr19_-_22182031 | 1.03 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr11_-_27455348 | 1.02 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr1_-_54593214 | 1.02 |
ENSDART00000152807
|
si:ch211-286b5.5
|
si:ch211-286b5.5 |
| chr22_-_13325475 | 1.02 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr8_-_27839244 | 0.99 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr23_-_24240728 | 0.99 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr14_-_9407573 | 0.98 |
ENSDART00000166739
|
NRK
|
si:zfos-2326c3.2 |
| chr1_+_25005968 | 0.96 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a (phosphatidylcholine-retinol O-acyltransferase) |
| chr6_-_43094926 | 0.93 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr13_-_31304614 | 0.92 |
|
|
|
| chr5_+_26925238 | 0.91 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr21_-_24552672 | 0.91 |
|
|
|
| chr20_-_48677794 | 0.90 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr13_+_16148867 | 0.90 |
ENSDART00000142408
|
anxa11a
|
annexin A11a |
| chr4_+_12616987 | 0.90 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr20_+_29307142 | 0.90 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_+_29307039 | 0.88 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_3970778 | 0.88 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr22_-_13018196 | 0.82 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr11_-_42934175 | 0.81 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_+_25794014 | 0.81 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr11_-_42933969 | 0.80 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr15_-_46670611 | 0.80 |
|
|
|
| chr9_-_41608298 | 0.80 |
|
|
|
| chr17_-_47893203 | 0.80 |
|
|
|
| chr13_-_44492897 | 0.79 |
|
|
|
| chr3_-_28120668 | 0.77 |
|
|
|
| chr13_+_42280697 | 0.77 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_-_44402452 | 0.77 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr22_+_25795022 | 0.76 |
ENSDART00000161120
|
vasna
|
vasorin a |
| chr4_+_12617165 | 0.75 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr2_-_50638153 | 0.75 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein-like 2b |
| chr23_+_28804842 | 0.74 |
ENSDART00000047378
|
sst3
|
somatostatin 3 |
| chr9_-_43736549 | 0.73 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr3_+_17387551 | 0.73 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr11_+_43924640 | 0.70 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr3_+_12132746 | 0.70 |
|
|
|
| chr12_+_27444832 | 0.69 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr11_+_19894390 | 0.67 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr22_-_12905648 | 0.65 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr11_-_42626842 | 0.64 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr4_+_11385828 | 0.64 |
|
|
|
| chr3_+_23607148 | 0.63 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr7_-_50359643 | 0.61 |
ENSDART00000174031
|
crtc3
|
CREB regulated transcription coactivator 3 |
| chr18_+_37034218 | 0.60 |
|
|
|
| chr3_-_30303173 | 0.59 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr13_+_18401965 | 0.58 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr2_+_29265967 | 0.58 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr6_+_11594333 | 0.57 |
ENSDART00000109552
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr7_-_50359583 | 0.57 |
ENSDART00000174031
|
crtc3
|
CREB regulated transcription coactivator 3 |
| chr9_-_39190534 | 0.56 |
|
|
|
| chr6_+_44891187 | 0.56 |
|
|
|
| chr6_+_16341787 | 0.54 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr9_-_41982635 | 0.54 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr25_+_6059393 | 0.53 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr6_+_44891278 | 0.51 |
|
|
|
| chr24_-_23155932 | 0.51 |
|
|
|
| chr8_+_6533379 | 0.50 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr12_-_17576472 | 0.50 |
ENSDART00000147282
|
pvalb9
|
parvalbumin 9 |
| chr18_-_43890514 | 0.49 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr6_-_40724581 | 0.47 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr25_-_21618526 | 0.46 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
dedicator of cytokinesis 4 |
| chr18_+_1353738 | 0.46 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr7_+_25793777 | 0.45 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr8_+_51061523 | 0.44 |
ENSDART00000166249
|
DISP3
|
dispatched RND transporter family member 3 |
| chr23_-_24240798 | 0.43 |
ENSDART00000135173
|
CR925709.1
|
ENSDARG00000092130 |
| KN149874v1_-_3544 | 0.42 |
ENSDART00000165981
|
CABZ01046348.1
|
ENSDARG00000099481 |
| chr8_+_30446599 | 0.42 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr23_+_17220363 | 0.41 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr11_+_19894420 | 0.41 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr11_-_6442836 | 0.40 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr14_-_6901415 | 0.40 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_+_31890795 | 0.39 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr7_+_30874387 | 0.39 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_+_33049445 | 0.37 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr3_-_8608620 | 0.37 |
|
|
|
| chr23_-_29138719 | 0.37 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr19_+_8693914 | 0.36 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr4_+_12613575 | 0.35 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr9_+_33049549 | 0.35 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr10_-_24422496 | 0.33 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr17_-_49355712 | 0.31 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr13_-_51409167 | 0.30 |
ENSDART00000174550
|
CABZ01090417.1
|
ENSDARG00000108080 |
| chr5_-_71277652 | 0.29 |
|
|
|
| chr3_-_33773025 | 0.27 |
|
|
|
| chr8_+_8889274 | 0.27 |
ENSDART00000142075
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr13_+_24453943 | 0.26 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr22_-_2942708 | 0.26 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr16_-_12282596 | 0.24 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr2_+_7127561 | 0.24 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr6_-_29094887 | 0.23 |
|
|
|
| chr2_+_29729161 | 0.23 |
ENSDART00000141666
|
AL845362.1
|
ENSDARG00000093205 |
| chr20_-_9448803 | 0.23 |
ENSDART00000133000
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr7_-_17527827 | 0.22 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr17_-_29885237 | 0.21 |
ENSDART00000009104
|
esrrga
|
estrogen-related receptor gamma a |
| chr13_+_25319230 | 0.20 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr11_+_19894502 | 0.20 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr23_-_36198253 | 0.19 |
|
|
|
| chr5_-_13872772 | 0.19 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr15_-_5732737 | 0.18 |
ENSDART00000109599
|
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr16_+_31890895 | 0.18 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr16_-_21973243 | 0.18 |
ENSDART00000039381
|
cops7a
|
COP9 constitutive photomorphogenic homolog subunit 7A |
| chr1_-_18581114 | 0.17 |
ENSDART00000178278
|
BX546494.1
|
ENSDARG00000107606 |
| chr2_-_27065203 | 0.17 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr1_+_9634016 | 0.16 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr4_-_15442709 | 0.15 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr18_+_1353857 | 0.14 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr17_-_45036982 | 0.12 |
ENSDART00000155043
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr24_-_14447136 | 0.12 |
|
|
|
| chr1_-_8439215 | 0.12 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr3_-_28120760 | 0.12 |
|
|
|
| chr9_+_41910191 | 0.12 |
|
|
|
| chr11_+_38013238 | 0.12 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr4_+_11465367 | 0.12 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr17_-_44446275 | 0.09 |
ENSDART00000165252
|
slc35f4
|
solute carrier family 35, member F4 |
| chr18_-_46279307 | 0.09 |
|
|
|
| chr18_-_36787879 | 0.08 |
|
|
|
| chr3_-_34431797 | 0.08 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr8_-_21110428 | 0.06 |
ENSDART00000135938
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr6_+_16342020 | 0.03 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr12_-_20493683 | 0.01 |
|
|
|
| chr20_+_54595334 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.7 | 2.2 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.4 | 1.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.4 | 1.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.4 | 2.5 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.3 | 1.2 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 2.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.3 | 2.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 2.2 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.2 | 1.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) protein oxidation(GO:0018158) |
| 0.2 | 0.7 | GO:0030431 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.2 | 0.6 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.2 | 1.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 1.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.7 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 5.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 3.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.5 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.9 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.0 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.3 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 2.1 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 1.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0030643 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.2 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.4 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 7.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 3.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 2.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 3.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.1 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 2.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |