Project
DANIO-CODE
Navigation
Downloads
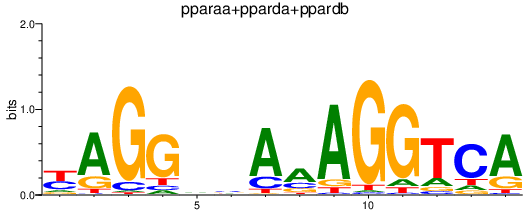
Results for pparaa+pparda+ppardb
Z-value: 0.71
Transcription factors associated with pparaa+pparda+ppardb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ppardb
|
ENSDARG00000009473 | peroxisome proliferator-activated receptor delta b |
|
pparaa
|
ENSDARG00000031777 | peroxisome proliferator-activated receptor alpha a |
|
pparda
|
ENSDARG00000044525 | peroxisome proliferator-activated receptor delta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pparda | dr10_dc_chr22_+_980951_980953 | -0.60 | 1.3e-02 | Click! |
| ppardb | dr10_dc_chr8_+_23682463_23682503 | 0.39 | 1.3e-01 | Click! |
| pparaa | dr10_dc_chr4_-_18660286_18660319 | 0.38 | 1.4e-01 | Click! |
Activity profile of pparaa+pparda+ppardb motif
Sorted Z-values of pparaa+pparda+ppardb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of pparaa+pparda+ppardb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_933677 | 6.03 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr1_+_25157675 | 2.63 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr25_+_21732255 | 2.56 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr14_-_49115338 | 1.65 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr2_-_43110857 | 1.63 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr13_-_37001997 | 1.62 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr4_-_16461748 | 1.58 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| KN149726v1_+_1094 | 1.50 |
|
|
|
| chr12_-_5085227 | 1.48 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr17_+_12254292 | 1.43 |
ENSDART00000160722
|
khk
|
ketohexokinase |
| chr25_+_5162045 | 1.39 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr25_+_22224346 | 1.36 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr4_-_24310846 | 1.21 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr5_+_37600633 | 1.19 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr13_+_28689749 | 1.14 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr19_+_41274734 | 1.14 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr23_+_44817648 | 1.12 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr23_+_2193582 | 1.12 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr6_-_30876091 | 1.10 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr10_-_44180588 | 1.00 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr1_+_40098861 | 0.91 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr20_-_53162473 | 0.88 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr17_+_443558 | 0.88 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr11_+_24720057 | 0.84 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr25_+_35797357 | 0.81 |
ENSDART00000152449
|
CR354435.1
|
ENSDARG00000039547 |
| chr8_-_12171880 | 0.79 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr10_+_4717800 | 0.79 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| KN149978v1_-_10669 | 0.78 |
ENSDART00000175723
|
CABZ01068178.1
|
ENSDARG00000105971 |
| chr22_+_38276148 | 0.75 |
ENSDART00000104504
|
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr17_-_16079935 | 0.73 |
|
|
|
| chr19_+_32570656 | 0.73 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr15_-_47264693 | 0.71 |
|
|
|
| chr12_-_31342432 | 0.68 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr15_+_895792 | 0.66 |
|
|
|
| chr23_-_3814291 | 0.66 |
|
|
|
| chr8_-_12171624 | 0.66 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr9_+_28301359 | 0.65 |
|
|
|
| chr25_+_17308788 | 0.60 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr20_+_15119519 | 0.60 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr7_-_23775835 | 0.59 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr5_+_18827478 | 0.59 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr1_+_11485480 | 0.57 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr12_-_7951265 | 0.57 |
|
|
|
| chr25_+_30701751 | 0.54 |
|
|
|
| chr21_-_18969970 | 0.51 |
ENSDART00000080269
|
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr11_+_11217547 | 0.51 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr7_-_33878076 | 0.49 |
ENSDART00000045374
|
smad3a
|
SMAD family member 3a |
| chr15_+_3296905 | 0.48 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr8_-_12171755 | 0.46 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr6_+_60060297 | 0.45 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr2_+_23600656 | 0.43 |
|
|
|
| chr2_-_39052185 | 0.42 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr10_-_41478085 | 0.42 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr20_+_15119754 | 0.42 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr22_-_17570275 | 0.42 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr12_-_34612758 | 0.41 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr9_-_41982635 | 0.41 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr16_-_50024848 | 0.41 |
ENSDART00000161782
|
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr16_+_28667291 | 0.39 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr15_-_47264619 | 0.38 |
|
|
|
| chr25_-_34608926 | 0.37 |
ENSDART00000130395
|
hist1h4l
|
histone 1, H4, like |
| chr6_-_39767452 | 0.36 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr17_+_25463178 | 0.36 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr12_+_27035744 | 0.35 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr16_+_5470419 | 0.35 |
|
|
|
| chr9_+_48521741 | 0.34 |
ENSDART00000145972
|
ccdc173
|
coiled-coil domain containing 173 |
| chr23_+_36241656 | 0.32 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr3_+_23621843 | 0.30 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr4_-_16461881 | 0.27 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| KN149947v1_-_2715 | 0.26 |
ENSDART00000158566
|
ENSDARG00000101294
|
ENSDARG00000101294 |
| chr9_+_28301214 | 0.26 |
|
|
|
| chr2_+_7341600 | 0.24 |
ENSDART00000146547
|
ENSDARG00000078299
|
ENSDARG00000078299 |
| chr22_+_25304162 | 0.24 |
ENSDART00000155836
ENSDART00000155006 |
si:ch211-226h8.15
|
si:ch211-226h8.15 |
| chr22_-_10744909 | 0.22 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr20_-_48773402 | 0.22 |
ENSDART00000161769
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr13_-_37002066 | 0.21 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr18_+_18465885 | 0.21 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr1_-_49274993 | 0.20 |
ENSDART00000050603
|
hadh
|
hydroxyacyl-CoA dehydrogenase |
| chr4_+_25868802 | 0.19 |
|
|
|
| chr18_+_16755239 | 0.19 |
ENSDART00000133490
|
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr12_+_26579286 | 0.19 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr15_+_494813 | 0.19 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr23_+_2193828 | 0.18 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr20_-_27038165 | 0.17 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr15_+_28249920 | 0.16 |
|
|
|
| chr23_-_24562107 | 0.16 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr8_+_8660457 | 0.15 |
ENSDART00000137780
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr8_-_24194821 | 0.14 |
ENSDART00000177464
|
CT025875.1
|
ENSDARG00000106046 |
| chr5_-_21520111 | 0.14 |
|
|
|
| chr10_-_41742329 | 0.13 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr15_+_17094877 | 0.11 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr22_+_34454253 | 0.11 |
ENSDART00000156615
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr6_+_28127514 | 0.10 |
|
|
|
| chr6_+_28127444 | 0.09 |
|
|
|
| chr18_+_12452 | 0.08 |
|
|
|
| chr21_-_1956120 | 0.05 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr3_+_58096720 | 0.04 |
ENSDART00000010395
ENSDART00000159755 ENSDART00000171149 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein IIa |
| chr11_+_24720206 | 0.03 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr15_-_29900 | 0.03 |
|
|
|
| chr15_+_28335293 | 0.02 |
ENSDART00000152536
|
myo1cb
|
myosin Ic, paralog b |
| chr24_-_11365575 | 0.01 |
ENSDART00000140217
|
prpf4bb
|
pre-mRNA processing factor 4Bb |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.4 | 1.1 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.3 | 2.6 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 1.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 2.1 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.2 | 1.6 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.7 | GO:1903793 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.6 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 0.5 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 1.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.9 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.9 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 1.2 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 0.9 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 8.8 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 1.0 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.2 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.5 | 1.4 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.3 | 2.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 1.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.1 | 0.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.6 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.2 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |