Project
DANIO-CODE
Navigation
Downloads
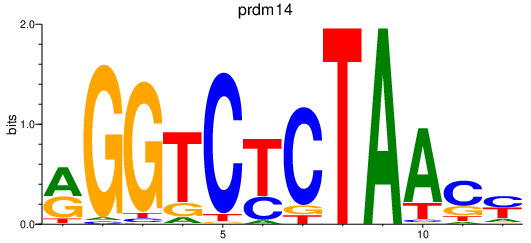
Results for prdm14
Z-value: 0.27
Transcription factors associated with prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm14
|
ENSDARG00000045371 | PR domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm14 | dr10_dc_chr24_+_14302079_14302118 | 0.55 | 2.7e-02 | Click! |
Activity profile of prdm14 motif
Sorted Z-values of prdm14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_24220413 | 0.46 |
ENSDART00000103176
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr20_+_22321303 | 0.37 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr13_-_7897936 | 0.34 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr15_+_24708696 | 0.32 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr4_+_29209923 | 0.26 |
ENSDART00000168709
|
ENSDARG00000104041
|
ENSDARG00000104041 |
| chr13_-_47767713 | 0.26 |
ENSDART00000045475
|
itga9
|
integrin, alpha 9 |
| chr2_-_32842855 | 0.25 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr5_-_29858748 | 0.22 |
|
|
|
| chr5_+_49093134 | 0.22 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr11_-_23079615 | 0.21 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr23_+_39665575 | 0.20 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr5_+_49093250 | 0.20 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr1_-_57842604 | 0.20 |
|
|
|
| chr7_+_48532749 | 0.19 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr4_+_50616647 | 0.19 |
|
|
|
| chr22_-_20317201 | 0.18 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
| chr6_-_19928053 | 0.17 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr1_+_41148042 | 0.17 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr14_-_30556788 | 0.16 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr7_-_32743153 | 0.16 |
|
|
|
| chr22_-_20316908 | 0.15 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr21_-_2116499 | 0.15 |
ENSDART00000167307
|
AL627305.3
|
ENSDARG00000102019 |
| chr5_+_27671064 | 0.14 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr2_+_32843206 | 0.13 |
ENSDART00000132191
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr18_+_48798941 | 0.12 |
ENSDART00000168937
|
bmp16
|
bone morphogenetic protein 16 |
| chr24_-_20987184 | 0.11 |
ENSDART00000010126
|
zdhhc23b
|
zinc finger, DHHC-type containing 23b |
| chr22_-_20317131 | 0.10 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
| chr16_-_31670211 | 0.10 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr13_+_21694608 | 0.09 |
|
|
|
| chr13_-_41235048 | 0.09 |
|
|
|
| chr22_-_20316792 | 0.08 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr25_-_35698109 | 0.08 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3b |
| chr13_+_21694465 | 0.06 |
|
|
|
| chr17_+_22359660 | 0.05 |
ENSDART00000162670
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr10_+_572931 | 0.05 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr13_-_31413855 | 0.05 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr2_-_32842678 | 0.05 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr9_+_33450418 | 0.05 |
ENSDART00000122803
|
usp9
|
ubiquitin specific peptidase 9 |
| chr22_-_9860697 | 0.04 |
ENSDART00000132304
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr5_+_62304435 | 0.04 |
ENSDART00000123243
|
TIMM22
|
translocase of inner mitochondrial membrane 22 |
| chr22_-_11106940 | 0.04 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr5_+_56885728 | 0.04 |
|
|
|
| chr9_-_40271416 | 0.04 |
|
|
|
| chr23_+_21334719 | 0.03 |
ENSDART00000141903
|
emc1
|
ER membrane protein complex subunit 1 |
| chr21_+_10699071 | 0.03 |
|
|
|
| chr6_-_39220951 | 0.03 |
ENSDART00000133305
ENSDART00000166290 |
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr11_+_13584479 | 0.03 |
ENSDART00000060251
|
wdr18
|
WD repeat domain 18 |
| chr7_-_33076034 | 0.02 |
|
|
|
| chr17_+_28657998 | 0.02 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr14_+_47022965 | 0.01 |
|
|
|
| chr9_+_39309903 | 0.00 |
|
|
|
| chr7_+_51049687 | 0.00 |
ENSDART00000114429
|
usp12b
|
ubiquitin specific peptidase 12b |
| chr13_+_21694572 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.2 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098594 | mucin granule(GO:0098594) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |