Project
DANIO-CODE
Navigation
Downloads
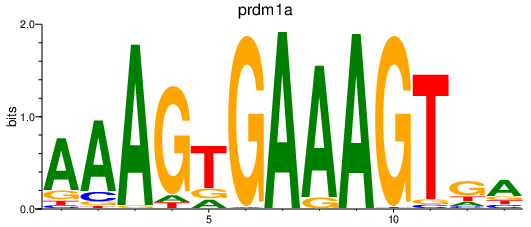
Results for prdm1a
Z-value: 0.92
Transcription factors associated with prdm1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm1a
|
ENSDARG00000002445 | PR domain containing 1a, with ZNF domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm1a | dr10_dc_chr16_-_7525980_7526051 | 0.80 | 2.3e-04 | Click! |
Activity profile of prdm1a motif
Sorted Z-values of prdm1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_40894631 | 2.69 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr24_-_39722595 | 2.51 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr22_+_33177432 | 2.46 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr15_-_35563367 | 2.46 |
|
|
|
| chr7_-_8635687 | 2.31 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr15_+_40707917 | 2.15 |
ENSDART00000154187
ENSDART00000042082 |
fat3a
|
FAT atypical cadherin 3a |
| chr7_-_35438739 | 2.12 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr11_+_21749658 | 2.10 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr6_-_39315024 | 2.10 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr1_-_44009572 | 1.97 |
ENSDART00000144900
|
ENSDARG00000079632
|
ENSDARG00000079632 |
| chr8_+_23716843 | 1.97 |
ENSDART00000136547
|
rpl10a
|
ribosomal protein L10a |
| chr22_+_30234718 | 1.87 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr23_+_2391223 | 1.84 |
|
|
|
| chr9_-_22140954 | 1.82 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr13_+_30781652 | 1.82 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr8_+_39964695 | 1.81 |
ENSDART00000073782
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr8_+_15968469 | 1.81 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr10_-_35036872 | 1.80 |
ENSDART00000170625
|
smad9
|
SMAD family member 9 |
| chr25_-_22890657 | 1.76 |
|
|
|
| chr16_+_42925950 | 1.75 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr21_-_20674965 | 1.65 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr6_+_17959219 | 1.61 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr6_-_58152436 | 1.55 |
ENSDART00000170752
|
tox2
|
TOX high mobility group box family member 2 |
| chr10_-_42032702 | 1.52 |
|
|
|
| chr18_-_15405161 | 1.50 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr12_+_32936230 | 1.47 |
ENSDART00000153146
|
rbfox3a
|
RNA binding protein, fox-1 homolog (C. elegans) 3a |
| chr25_+_222244 | 1.45 |
ENSDART00000155344
|
ENSDARG00000073905
|
ENSDARG00000073905 |
| chr20_-_20502615 | 1.45 |
|
|
|
| chr17_+_1058137 | 1.45 |
|
|
|
| chr6_-_40660116 | 1.44 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr19_-_47366517 | 1.41 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr3_+_16826320 | 1.39 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr13_+_30781718 | 1.36 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr4_+_15870605 | 1.36 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr5_-_43334777 | 1.35 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr11_-_18542803 | 1.26 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr2_-_43997672 | 1.25 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr8_-_50270783 | 1.24 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr16_-_43441084 | 1.21 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr23_+_6298911 | 1.20 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr17_-_31041977 | 1.18 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr16_+_5508950 | 1.17 |
ENSDART00000138674
|
plecb
|
plectin b |
| chr14_-_12531063 | 1.16 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr5_-_63531729 | 1.15 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr12_+_5154886 | 1.14 |
ENSDART00000163897
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr4_+_5124186 | 1.09 |
ENSDART00000127874
|
ccnd2b
|
cyclin D2, b |
| chr3_-_23107763 | 1.07 |
ENSDART00000154058
|
AL929222.2
|
ENSDARG00000097738 |
| chr19_+_9113356 | 1.04 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr7_-_68229145 | 1.03 |
|
|
|
| chr5_-_63422783 | 1.01 |
ENSDART00000083684
|
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr1_+_30974088 | 1.00 |
ENSDART00000144957
|
as3mt
|
arsenite methyltransferase |
| chr19_+_48582114 | 0.99 |
ENSDART00000157424
|
CU693379.1
|
ENSDARG00000098777 |
| chr11_-_34367105 | 0.98 |
ENSDART00000157625
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr15_-_39756101 | 0.96 |
|
|
|
| chr1_+_44009649 | 0.94 |
ENSDART00000131296
|
ssrp1b
|
structure specific recognition protein 1b |
| chr8_+_50953220 | 0.93 |
ENSDART00000124748
|
b2ml
|
beta-2-microglobulin, like |
| chr7_+_48597712 | 0.91 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr13_-_1021000 | 0.91 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr11_-_25803101 | 0.91 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr12_-_37124892 | 0.90 |
ENSDART00000146142
|
pmp22b
|
peripheral myelin protein 22b |
| chr14_+_20862466 | 0.89 |
ENSDART00000059796
|
ENSDARG00000019213
|
ENSDARG00000019213 |
| chr7_-_27877036 | 0.89 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr8_+_39525254 | 0.89 |
|
|
|
| chr7_+_65532390 | 0.89 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr23_-_27196543 | 0.86 |
ENSDART00000023218
ENSDART00000168175 ENSDART00000059346 |
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr6_-_39315608 | 0.86 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr10_+_10252074 | 0.84 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr23_-_28286971 | 0.82 |
|
|
|
| chr25_+_6858286 | 0.82 |
|
|
|
| chr5_-_46382728 | 0.82 |
|
|
|
| chr11_+_21749918 | 0.81 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr24_+_37266599 | 0.80 |
ENSDART00000140412
|
si:dkey-224b4.5
|
si:dkey-224b4.5 |
| chr11_-_38225034 | 0.80 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr3_-_12087518 | 0.80 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr10_-_37098396 | 0.80 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr5_-_43686611 | 0.78 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr2_-_16586136 | 0.77 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_31210417 | 0.77 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr23_-_3760604 | 0.77 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr7_-_8635989 | 0.76 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr10_-_29930133 | 0.76 |
ENSDART00000055913
|
hist2h2l
|
histone 2, H2, like |
| chr7_+_37105282 | 0.75 |
|
|
|
| chr7_+_26980284 | 0.75 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr16_+_13964949 | 0.74 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr25_-_23648033 | 0.71 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr9_-_8475669 | 0.68 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr23_+_24778062 | 0.67 |
|
|
|
| chr13_+_41885396 | 0.67 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr15_-_1042743 | 0.67 |
ENSDART00000154195
|
si:dkey-77f5.6
|
si:dkey-77f5.6 |
| chr20_-_47576935 | 0.66 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr11_-_16067646 | 0.66 |
|
|
|
| chr13_+_41885308 | 0.64 |
ENSDART00000126013
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr7_-_68229018 | 0.64 |
|
|
|
| chr9_-_19951894 | 0.64 |
ENSDART00000146841
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr18_-_14723138 | 0.64 |
ENSDART00000010129
|
pdf
|
peptide deformylase (mitochondrial) |
| chr22_-_23641813 | 0.64 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr16_-_49282668 | 0.62 |
|
|
|
| chr14_-_4014025 | 0.61 |
ENSDART00000157782
|
irf2
|
interferon regulatory factor 2 |
| chr18_-_18554151 | 0.61 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr11_-_1792616 | 0.60 |
|
|
|
| chr3_-_34417082 | 0.59 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr10_-_210681 | 0.58 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr18_+_14725148 | 0.56 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr8_+_15968636 | 0.56 |
ENSDART00000141173
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_22470384 | 0.56 |
ENSDART00000125805
|
si:ch211-261n11.8
|
si:ch211-261n11.8 |
| chr4_+_17535295 | 0.55 |
ENSDART00000172169
ENSDART00000021610 |
recql
|
RecQ helicase-like |
| chr8_-_14338087 | 0.55 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr11_-_38225208 | 0.55 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr17_+_860032 | 0.55 |
ENSDART00000160400
|
gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr14_-_46645127 | 0.52 |
|
|
|
| chr10_+_7043530 | 0.52 |
|
|
|
| chr4_-_9666436 | 0.51 |
ENSDART00000133214
|
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr12_-_16331781 | 0.51 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr24_+_17056023 | 0.51 |
ENSDART00000149225
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr18_+_50279433 | 0.49 |
ENSDART00000143911
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr20_+_4750766 | 0.49 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr6_-_39313420 | 0.49 |
|
|
|
| chr4_-_74789596 | 0.48 |
ENSDART00000174356
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr9_-_22141138 | 0.47 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr8_-_14014576 | 0.46 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr20_-_51585893 | 0.46 |
ENSDART00000151426
|
si:ch73-91k6.2
|
si:ch73-91k6.2 |
| chr8_-_27839244 | 0.45 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr5_-_42304154 | 0.44 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr4_+_17535561 | 0.42 |
ENSDART00000101198
|
recql
|
RecQ helicase-like |
| chr11_-_38280444 | 0.42 |
|
|
|
| chr25_-_7585373 | 0.42 |
ENSDART00000157076
|
prdm11
|
PR domain containing 11 |
| chr23_+_35405877 | 0.42 |
ENSDART00000159218
|
acp1
|
acid phosphatase 1, soluble |
| chr13_-_49921086 | 0.41 |
ENSDART00000165066
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr10_+_10252239 | 0.40 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr7_+_37105966 | 0.40 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr22_-_23721742 | 0.39 |
ENSDART00000163228
|
cfh
|
complement factor H |
| chr5_-_14990023 | 0.39 |
|
|
|
| chr23_+_24778090 | 0.38 |
|
|
|
| chr14_+_21388983 | 0.37 |
ENSDART00000137589
|
ran
|
RAN, member RAS oncogene family |
| chr19_-_32900108 | 0.35 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr23_+_2391317 | 0.35 |
|
|
|
| chr25_-_23648222 | 0.33 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr6_+_24561921 | 0.33 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr7_+_65532771 | 0.33 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr22_+_2253153 | 0.32 |
ENSDART00000133751
|
AL929017.2
|
ENSDARG00000095449 |
| chr11_-_38280529 | 0.32 |
|
|
|
| chr16_+_7874573 | 0.31 |
|
|
|
| chr5_+_58824344 | 0.29 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr3_-_19847002 | 0.29 |
|
|
|
| chr19_+_8693914 | 0.28 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr14_-_46645261 | 0.28 |
|
|
|
| chr7_+_65532871 | 0.26 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr10_+_28700844 | 0.25 |
ENSDART00000047776
|
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr8_+_39964882 | 0.24 |
ENSDART00000134452
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr14_+_10795333 | 0.23 |
|
|
|
| chr11_-_34366745 | 0.23 |
ENSDART00000157625
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr4_-_22617898 | 0.22 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr9_+_8475950 | 0.21 |
|
|
|
| chr18_+_50279394 | 0.21 |
ENSDART00000143911
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr19_-_35642784 | 0.21 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr11_-_44223754 | 0.20 |
ENSDART00000172828
|
mfn1b
|
mitofusin 1b |
| chr4_+_22617563 | 0.20 |
|
|
|
| chr9_-_15236879 | 0.17 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr25_-_29914241 | 0.17 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr4_+_672670 | 0.16 |
ENSDART00000169748
|
fbxo7
|
F-box protein 7 |
| chr20_-_27038165 | 0.16 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr9_-_32803650 | 0.16 |
|
|
|
| chr23_-_7048903 | 0.15 |
ENSDART00000149656
|
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr24_+_24777962 | 0.14 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr7_-_32511304 | 0.13 |
ENSDART00000052497
|
gas2b
|
growth arrest-specific 2b |
| chr1_-_18084246 | 0.12 |
|
|
|
| chr3_-_41369324 | 0.12 |
|
|
|
| chr2_+_42155813 | 0.12 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr10_-_31334198 | 0.09 |
ENSDART00000134866
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr13_-_49920975 | 0.09 |
ENSDART00000165066
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr24_+_9740818 | 0.06 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr13_-_43503505 | 0.06 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr5_-_41044841 | 0.06 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr7_-_37292692 | 0.05 |
ENSDART00000149052
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr22_-_8479330 | 0.05 |
ENSDART00000140146
ENSDART00000136891 |
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr25_-_22242337 | 0.02 |
|
|
|
| chr19_+_7196277 | 0.02 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr4_-_6365088 | 0.01 |
ENSDART00000140100
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr19_+_12430505 | 0.00 |
ENSDART00000052240
|
psmg2
|
proteasome (prosome, macropain) assembly chaperone 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 1.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.4 | 1.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.3 | 2.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 3.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.2 | 1.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.2 | 0.4 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.2 | 0.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 0.6 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 1.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.4 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 2.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.7 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.9 | GO:0042102 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 3.0 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.4 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.8 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.1 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 2.3 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.0 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.2 | GO:0009416 | response to light stimulus(GO:0009416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 1.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 1.0 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 1.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 16.4 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |