Project
DANIO-CODE
Navigation
Downloads
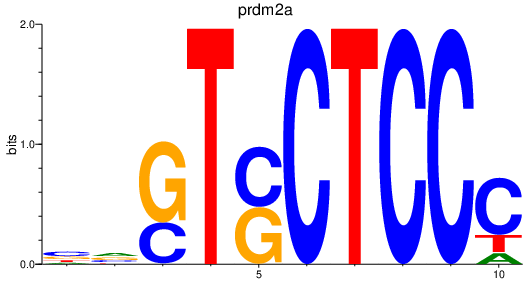
Results for prdm2a
Z-value: 5.70
Transcription factors associated with prdm2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm2a
|
ENSDARG00000090721 | PR domain containing 2, with ZNF domain a |
Activity profile of prdm2a motif
Sorted Z-values of prdm2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_24310846 | 14.08 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr7_+_39173765 | 13.86 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr6_+_33556031 | 13.48 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr20_+_31366832 | 11.59 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr23_-_9556986 | 11.57 |
ENSDART00000139688
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr13_-_49826330 | 10.95 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr15_-_2677200 | 10.92 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr23_-_35595271 | 10.92 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr14_+_20809272 | 10.77 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr13_-_18704185 | 10.72 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr21_-_1716495 | 10.24 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr6_-_43094573 | 10.06 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr5_+_59845054 | 10.02 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr5_+_59844830 | 9.66 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr15_-_9026081 | 9.66 |
ENSDART00000126708
|
rhoub
|
ras homolog family member Ub |
| chr3_-_6078015 | 9.64 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr18_+_5899355 | 9.37 |
|
|
|
| chr18_+_6551983 | 9.31 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr17_+_28690237 | 8.96 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr6_+_33556155 | 8.95 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr2_+_24881103 | 8.88 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr15_-_2689005 | 8.71 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr22_+_37944019 | 8.69 |
|
|
|
| chr7_-_28425307 | 8.56 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr13_-_7434672 | 8.49 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr14_+_35885903 | 8.44 |
ENSDART00000052569
|
pitx2
|
paired-like homeodomain 2 |
| chr9_-_3429153 | 8.42 |
ENSDART00000111386
|
dlx2a
|
distal-less homeobox 2a |
| chr14_-_46359062 | 8.39 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr23_-_27226280 | 8.36 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr17_-_49878964 | 8.32 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr14_+_34683602 | 8.18 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr7_+_6814828 | 7.92 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr2_-_39710140 | 7.81 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr6_+_55022668 | 7.77 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr10_-_26468280 | 7.76 |
ENSDART00000128894
|
dchs1b
|
dachsous cadherin-related 1b |
| chr25_-_10992022 | 7.74 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr3_-_54866453 | 7.70 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr4_+_8531280 | 7.70 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr12_+_32936230 | 7.61 |
ENSDART00000153146
|
rbfox3a
|
RNA binding protein, fox-1 homolog (C. elegans) 3a |
| chr1_+_50277442 | 7.61 |
ENSDART00000150420
|
otx1a
|
orthodenticle homeobox 1a |
| chr19_-_35739239 | 7.58 |
|
|
|
| KN149968v1_+_15749 | 7.56 |
ENSDART00000168977
|
ENSDARG00000102503
|
ENSDARG00000102503 |
| chr7_-_8635687 | 7.49 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr8_-_30233470 | 7.48 |
ENSDART00000139864
|
zgc:162939
|
zgc:162939 |
| chr23_+_42381296 | 7.46 |
ENSDART00000158684
ENSDART00000159985 ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr15_+_38957922 | 7.40 |
ENSDART00000141086
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr4_-_16556116 | 7.36 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr10_-_11806373 | 7.36 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr9_+_53788118 | 7.35 |
ENSDART00000125715
ENSDART00000166582 ENSDART00000162224 |
dct
|
dopachrome tautomerase |
| chr11_-_4216204 | 7.33 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr23_-_18958008 | 7.33 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr23_-_29138952 | 7.26 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr20_+_4222097 | 7.21 |
ENSDART00000097543
|
im:7142702
|
im:7142702 |
| chr9_+_26292183 | 7.14 |
|
|
|
| chr7_+_22552724 | 7.14 |
ENSDART00000101459
ENSDART00000159743 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr5_+_53938629 | 7.13 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr3_-_30554400 | 7.10 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr10_-_25695574 | 6.97 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr13_+_28601627 | 6.92 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr16_+_53238110 | 6.87 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr6_-_343209 | 6.81 |
|
|
|
| chr19_+_7234029 | 6.77 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr1_-_48853800 | 6.77 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr10_+_9052152 | 6.75 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr23_-_4990023 | 6.71 |
ENSDART00000142699
|
taz
|
tafazzin |
| chr13_-_43027245 | 6.57 |
ENSDART00000099601
|
ENSDARG00000068784
|
ENSDARG00000068784 |
| chr9_+_54692834 | 6.53 |
|
|
|
| chr10_-_36462538 | 6.53 |
ENSDART00000161823
|
ubl3a
|
ubiquitin-like 3a |
| chr24_-_9151388 | 6.53 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr13_+_41998500 | 6.53 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr14_-_4166292 | 6.49 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr8_+_28584427 | 6.47 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr2_-_59531831 | 6.43 |
ENSDART00000168568
|
CU861477.1
|
ENSDARG00000100128 |
| chr16_-_17164459 | 6.40 |
ENSDART00000178443
|
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr10_+_3728747 | 6.39 |
|
|
|
| chr25_+_35901081 | 6.37 |
ENSDART00000042271
ENSDART00000158027 |
irx3b
|
iroquois homeobox 3b |
| chr6_-_12487617 | 6.35 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr3_+_31542311 | 6.32 |
|
|
|
| chr20_+_54512601 | 6.31 |
ENSDART00000169386
|
faua
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a |
| chr4_+_7668939 | 6.26 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr16_-_31763183 | 6.25 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr24_-_21114505 | 6.21 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr2_-_30340646 | 6.17 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr6_-_19928053 | 6.15 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr9_-_32942783 | 6.08 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr8_+_15989815 | 5.99 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| KN149932v1_+_27584 | 5.95 |
|
|
|
| chr5_+_37053530 | 5.92 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr25_-_4376129 | 5.92 |
|
|
|
| chr7_+_58718614 | 5.91 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr3_+_22412059 | 5.90 |
|
|
|
| chr23_-_27226387 | 5.87 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr12_+_689413 | 5.85 |
ENSDART00000174804
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr16_+_54906517 | 5.85 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| chr20_-_9212177 | 5.84 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr25_-_36760720 | 5.83 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr22_-_22694468 | 5.82 |
ENSDART00000166794
|
nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr25_+_30746445 | 5.79 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr20_-_6822668 | 5.75 |
ENSDART00000169569
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr10_-_32580145 | 5.75 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr2_-_816669 | 5.72 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr5_-_36349284 | 5.70 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr14_-_2008649 | 5.65 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr3_-_7769397 | 5.61 |
|
|
|
| chr3_-_35928594 | 5.60 |
|
|
|
| chr8_+_26103807 | 5.59 |
|
|
|
| chr9_+_46038777 | 5.59 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr16_-_53896201 | 5.56 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr10_+_10252074 | 5.53 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr7_-_32888309 | 5.53 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr8_-_4451417 | 5.50 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr1_-_4696894 | 5.50 |
ENSDART00000103724
|
ndfip2
|
Nedd4 family interacting protein 2 |
| chr25_-_33670713 | 5.49 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr21_-_43020159 | 5.47 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr22_-_17652938 | 5.43 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr21_-_9708608 | 5.42 |
|
|
|
| chr20_-_21773202 | 5.41 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr4_+_11312432 | 5.39 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr19_-_29205158 | 5.39 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr18_+_7607055 | 5.38 |
ENSDART00000140784
|
CR318588.4
|
ENSDARG00000095556 |
| chr4_+_14972708 | 5.38 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr7_-_34656224 | 5.37 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3a |
| chr12_+_11042472 | 5.36 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr14_-_31128891 | 5.34 |
|
|
|
| chr15_-_14616083 | 5.34 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr1_+_1953714 | 5.31 |
ENSDART00000164488
ENSDART00000167050 ENSDART00000122626 ENSDART00000128187 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr2_+_3701942 | 5.30 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr22_+_5697877 | 5.26 |
ENSDART00000063484
|
si:dkey-222f2.1
|
si:dkey-222f2.1 |
| chr2_-_35584440 | 5.25 |
ENSDART00000125298
|
tnw
|
tenascin W |
| chr3_+_59560881 | 5.24 |
ENSDART00000084738
|
ppp1r27a
|
protein phosphatase 1, regulatory subunit 27a |
| chr14_+_41805073 | 5.24 |
ENSDART00000074362
|
pcdh18b
|
protocadherin 18b |
| chr2_+_49837788 | 5.22 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr20_+_34640226 | 5.21 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr2_-_49114158 | 5.19 |
|
|
|
| chr9_-_30243398 | 5.19 |
ENSDART00000139863
ENSDART00000140040 |
dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr18_-_45892 | 5.17 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr23_-_41690232 | 5.16 |
|
|
|
| chr7_-_56493164 | 5.16 |
|
|
|
| chr5_+_62987426 | 5.15 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr23_+_42449144 | 5.14 |
ENSDART00000171119
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr19_-_23037220 | 5.12 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr14_-_16023198 | 5.12 |
|
|
|
| chr3_+_12287067 | 5.10 |
ENSDART00000158060
|
vasnb
|
vasorin b |
| chr16_+_5625301 | 5.07 |
|
|
|
| chr14_+_34683695 | 5.07 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr20_-_54742983 | 5.04 |
|
|
|
| chr2_+_33399405 | 5.04 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_-_23587330 | 5.02 |
ENSDART00000167246
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr15_+_14918849 | 5.02 |
ENSDART00000164119
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr20_+_7337738 | 5.01 |
ENSDART00000165596
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr19_-_35512932 | 4.97 |
|
|
|
| chr14_-_46359248 | 4.97 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr2_+_26523457 | 4.96 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr18_-_45750 | 4.96 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_+_24476213 | 4.94 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr20_-_26568204 | 4.93 |
ENSDART00000158213
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr16_-_13704905 | 4.92 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr9_+_17341042 | 4.92 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr19_-_35513216 | 4.91 |
|
|
|
| chr11_-_471166 | 4.88 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr12_+_27370834 | 4.88 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr6_-_27901354 | 4.83 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr3_-_30554490 | 4.81 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr10_-_32580373 | 4.79 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr3_-_55995924 | 4.78 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr6_-_49674729 | 4.77 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr22_+_24362712 | 4.74 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr12_+_41016238 | 4.74 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| KN149955v1_+_4206 | 4.73 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr20_+_26803282 | 4.72 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr23_+_24711233 | 4.71 |
|
|
|
| chr14_+_4810147 | 4.71 |
ENSDART00000114678
|
nanos1
|
nanos homolog 1 |
| chr24_-_26165778 | 4.71 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr16_+_29060022 | 4.69 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr24_-_14567403 | 4.69 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| KN149955v1_+_4134 | 4.67 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr25_+_24152717 | 4.64 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr19_+_14197118 | 4.62 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr19_+_1797470 | 4.62 |
|
|
|
| chr2_+_49683715 | 4.60 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr10_-_10374032 | 4.57 |
ENSDART00000134929
|
BX323065.1
|
ENSDARG00000095365 |
| chr20_-_11111340 | 4.56 |
|
|
|
| chr4_+_21166170 | 4.53 |
ENSDART00000036886
|
nav3
|
neuron navigator 3 |
| chr3_-_10342940 | 4.51 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr12_+_18403055 | 4.50 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr6_-_12953515 | 4.50 |
ENSDART00000150036
ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr4_-_19027117 | 4.49 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr23_+_18176720 | 4.46 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr10_+_33462953 | 4.46 |
ENSDART00000137089
|
ryr1a
|
ryanodine receptor 1a (skeletal) |
| chr10_-_1799784 | 4.46 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_+_24059652 | 4.44 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr12_+_30448812 | 4.39 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr16_-_12579115 | 4.38 |
ENSDART00000147483
|
ephb6
|
eph receptor B6 |
| chr23_+_2193582 | 4.36 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr25_+_15551474 | 4.34 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr8_-_19311772 | 4.33 |
ENSDART00000138881
ENSDART00000010604 |
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr24_-_24001766 | 4.29 |
ENSDART00000173052
ENSDART00000173220 |
map7d2b
|
MAP7 domain containing 2b |
| chr11_-_27260783 | 4.28 |
|
|
|
| chr6_-_14010044 | 4.28 |
ENSDART00000149177
ENSDART00000174839 ENSDART00000089577 |
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr21_+_21337628 | 4.27 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 3.9 | GO:0034369 | protein-lipid complex remodeling(GO:0034368) plasma lipoprotein particle remodeling(GO:0034369) |
| 3.7 | 11.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 3.5 | 10.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 3.5 | 10.5 | GO:0008306 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 3.5 | 10.4 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 3.4 | 10.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 3.1 | 21.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 2.9 | 8.8 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 2.8 | 8.4 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 2.1 | 8.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 2.0 | 6.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.9 | 7.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 1.8 | 7.3 | GO:0048103 | cell proliferation in forebrain(GO:0021846) neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 1.8 | 5.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 1.8 | 5.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 1.8 | 7.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.7 | 9.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 1.6 | 12.4 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 1.5 | 22.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 1.5 | 3.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.4 | 10.1 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.4 | 5.7 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 1.4 | 4.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 1.3 | 10.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.3 | 3.9 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 1.3 | 7.8 | GO:0007343 | egg activation(GO:0007343) |
| 1.3 | 11.6 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 1.3 | 6.4 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 1.3 | 7.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 1.2 | 3.7 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 1.2 | 3.7 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.2 | 11.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 1.2 | 4.9 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 1.2 | 8.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 1.1 | 13.7 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 1.1 | 6.7 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 1.1 | 3.3 | GO:1903010 | regulation of bone development(GO:1903010) |
| 1.1 | 5.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 1.1 | 7.7 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 1.1 | 5.4 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 1.1 | 3.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 1.0 | 5.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 1.0 | 6.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.0 | 4.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.9 | 11.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.9 | 11.0 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 4.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.9 | 0.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.8 | 6.7 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.8 | 19.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.7 | 20.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.7 | 6.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.7 | 2.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.7 | 2.7 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.7 | 3.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 13.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.7 | 5.9 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 15.3 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.6 | 3.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.6 | 9.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.6 | 3.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.6 | 1.8 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.6 | 3.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.6 | 6.9 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.6 | 4.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.6 | 3.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.6 | 10.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 6.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.6 | 6.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.6 | 10.1 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.6 | 7.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.6 | 5.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.6 | 3.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 4.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 5.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.5 | 8.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.5 | 2.5 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.5 | 8.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.5 | 2.9 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) |
| 0.5 | 5.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 2.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 3.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.4 | 4.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 2.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.4 | 8.2 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.4 | 3.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.4 | 6.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.4 | 7.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.4 | 1.7 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.4 | 7.1 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.4 | 9.3 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.4 | 3.1 | GO:0097028 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.4 | 2.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 1.1 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.4 | 2.6 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.4 | 7.8 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.4 | 4.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.4 | 6.9 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.3 | 2.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.3 | 1.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.3 | 1.0 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 6.4 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.3 | 3.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 2.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 2.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 3.6 | GO:0031581 | cell-substrate junction assembly(GO:0007044) hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.3 | 3.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 | 5.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.3 | 1.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.3 | 3.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.3 | 5.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.3 | 1.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 18.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.3 | 1.3 | GO:0002551 | mast cell chemotaxis(GO:0002551) positive regulation of leukocyte chemotaxis(GO:0002690) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.3 | 8.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.2 | 5.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.2 | 9.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 1.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 3.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 2.0 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.2 | 3.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.2 | 7.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.2 | 8.3 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.2 | 3.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 3.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 22.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 5.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 1.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 6.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.2 | 2.6 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.2 | 5.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.2 | 4.7 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.2 | 6.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.2 | 4.7 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.2 | 0.7 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 12.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 5.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.2 | 2.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 1.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 11.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 7.7 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 0.8 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 2.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 3.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.5 | GO:0043627 | response to estrogen(GO:0043627) cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 3.7 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 6.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 2.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.6 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 5.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 3.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 3.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 3.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 8.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 2.8 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.1 | 0.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 1.6 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.1 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 2.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 1.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 7.1 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 3.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 2.1 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 22.5 | GO:0055085 | transmembrane transport(GO:0055085) |
| 0.0 | 3.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 7.6 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 4.3 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.7 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 1.1 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 6.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 3.3 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 1.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 5.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.3 | 11.6 | GO:0042627 | chylomicron(GO:0042627) |
| 1.7 | 8.3 | GO:0008091 | spectrin(GO:0008091) |
| 1.4 | 5.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 1.2 | 7.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.1 | 10.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 1.0 | 12.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 1.0 | 5.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.9 | 6.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.9 | 2.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.7 | 4.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.6 | 13.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 5.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 4.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 4.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 1.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 22.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 8.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 3.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 26.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.3 | 11.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 7.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 8.9 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.3 | 7.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 2.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 3.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 5.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 8.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 26.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 7.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 8.0 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 6.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 6.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 71.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 9.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 15.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 64.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 24.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 9.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 10.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.9 | GO:0005740 | mitochondrial envelope(GO:0005740) mitochondrial membrane(GO:0031966) |
| 0.0 | 4.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 3.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 22.4 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 3.3 | 9.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 2.0 | 5.9 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 1.9 | 9.5 | GO:0005035 | death receptor activity(GO:0005035) |
| 1.8 | 7.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 1.7 | 13.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.7 | 6.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.7 | 6.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.7 | 11.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 1.5 | 6.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.4 | 10.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 1.4 | 6.9 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.3 | 5.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.3 | 10.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.3 | 7.8 | GO:0016918 | retinal binding(GO:0016918) |
| 1.2 | 4.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 1.1 | 11.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.0 | 5.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.9 | 6.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.9 | 3.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.9 | 7.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.8 | 3.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.8 | 4.2 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.8 | 2.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.8 | 5.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.8 | 3.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.7 | 4.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 3.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.7 | 7.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 22.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.7 | 7.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.7 | 6.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.7 | 5.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.7 | 3.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.7 | 5.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 3.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 3.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 3.0 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 3.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.5 | 4.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 24.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 5.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 17.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 10.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 9.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.5 | 18.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.5 | 1.4 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.4 | 2.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 10.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.4 | 5.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 6.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.3 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 4.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 6.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 5.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 23.3 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 5.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.3 | 5.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 3.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.3 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 2.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 4.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 4.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 3.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 1.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 7.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 1.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 5.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 1.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 4.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 5.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 5.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 11.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 72.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.9 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 13.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 2.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 11.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 20.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.8 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 0.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 13.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 9.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0016530 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.0 | 43.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 6.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 11.3 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 2.7 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 4.7 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 14.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 8.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 5.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 13.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 5.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 21.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 19.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 1.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 2.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 3.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 1.2 | 8.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.0 | 5.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.8 | 5.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 10.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.7 | 9.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.6 | 5.8 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.6 | 10.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.6 | 12.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.6 | 3.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 12.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.5 | 3.4 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.5 | 2.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 3.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 12.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 3.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 3.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 2.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 7.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 5.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 8.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 7.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 6.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 11.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 2.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |