Project
DANIO-CODE
Navigation
Downloads
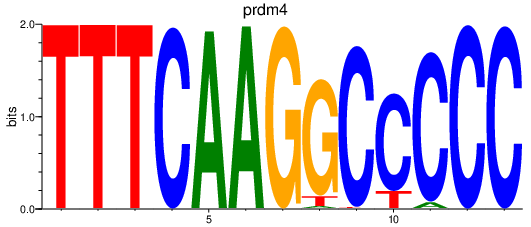
Results for prdm4
Z-value: 0.70
Transcription factors associated with prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm4
|
ENSDARG00000017366 | PR domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm4 | dr10_dc_chr4_-_14927871_14927908 | 0.64 | 7.7e-03 | Click! |
Activity profile of prdm4 motif
Sorted Z-values of prdm4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_31490043 | 3.70 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr9_-_13383818 | 3.59 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr23_-_24755654 | 3.00 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr22_-_21021645 | 2.10 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr2_-_41873625 | 1.36 |
ENSDART00000155577
|
ENSDARG00000056490
|
ENSDARG00000056490 |
| chr9_-_13383708 | 0.98 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr3_-_56969897 | 0.88 |
|
|
|
| chr20_-_42306410 | 0.76 |
ENSDART00000074959
|
slc35f1
|
solute carrier family 35, member F1 |
| chr12_+_34615991 | 0.69 |
|
|
|
| chr24_-_16994956 | 0.63 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr4_+_45204456 | 0.55 |
ENSDART00000158744
|
BX927336.3
|
ENSDARG00000102848 |
| chr17_-_31594647 | 0.51 |
ENSDART00000135468
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr6_-_9471897 | 0.47 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr3_+_32293441 | 0.42 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr22_-_21021866 | 0.36 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr19_+_30212565 | 0.24 |
|
|
|
| chr23_-_31474216 | 0.20 |
ENSDART00000132736
|
lca5
|
Leber congenital amaurosis 5 |
| chr2_+_41973443 | 0.04 |
|
|
|
| chr20_-_26102193 | 0.03 |
ENSDART00000136518
|
capn3b
|
calpain 3b |
| chr3_+_40265548 | 0.01 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 3.7 | GO:0003306 | Wnt signaling pathway involved in heart development(GO:0003306) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 5.0 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |