Project
DANIO-CODE
Navigation
Downloads
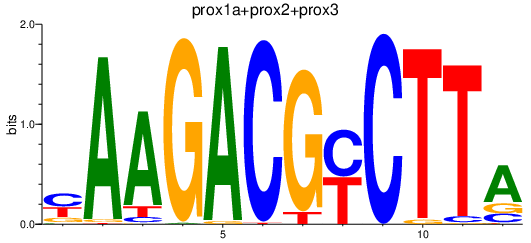
Results for prox1a+prox2+prox3
Z-value: 1.23
Transcription factors associated with prox1a+prox2+prox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prox2
|
ENSDARG00000041952 | prospero homeobox 2 |
|
prox1a
|
ENSDARG00000055158 | prospero homeobox 1a |
|
prox3
|
ENSDARG00000088810 | prospero homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prox1a | dr10_dc_chr17_-_32913432_32913509 | -0.85 | 3.2e-05 | Click! |
| prox1b | dr10_dc_chr7_-_19688306_19688428 | 0.35 | 1.9e-01 | Click! |
Activity profile of prox1a+prox2+prox3 motif
Sorted Z-values of prox1a+prox2+prox3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of prox1a+prox2+prox3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_32747592 | 5.10 |
|
|
|
| chr14_+_24543399 | 3.34 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr24_+_40321763 | 3.24 |
|
|
|
| chr5_+_22006830 | 2.60 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr19_+_20404995 | 2.48 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr15_-_14102102 | 2.33 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr16_-_55211053 | 2.20 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr7_-_44265768 | 2.12 |
ENSDART00000149072
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr23_+_25952724 | 2.08 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_-_8801877 | 2.07 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr9_+_8990576 | 2.02 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr19_+_20405107 | 1.96 |
ENSDART00000151066
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr9_+_8990774 | 1.95 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr14_+_28167213 | 1.93 |
ENSDART00000157306
ENSDART00000166375 ENSDART00000167439 ENSDART00000017075 |
xiap
|
X-linked inhibitor of apoptosis |
| chr1_-_12575687 | 1.92 |
|
|
|
| chr15_-_29229170 | 1.85 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr16_+_4353561 | 1.83 |
ENSDART00000060729
|
yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr8_+_11287550 | 1.78 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr3_-_26060787 | 1.76 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr6_-_32424559 | 1.76 |
ENSDART00000151002
|
usp1
|
ubiquitin specific peptidase 1 |
| chr6_-_31798839 | 1.75 |
|
|
|
| chr12_+_6032235 | 1.68 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr3_-_21006698 | 1.62 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr14_+_28167358 | 1.61 |
ENSDART00000157306
ENSDART00000166375 ENSDART00000167439 ENSDART00000017075 |
xiap
|
X-linked inhibitor of apoptosis |
| chr17_+_15666078 | 1.60 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr3_-_5523316 | 1.59 |
ENSDART00000159308
|
trim35-7
|
tripartite motif containing 35-7 |
| chr13_-_25711537 | 1.52 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr4_+_5187879 | 1.52 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr21_-_32747544 | 1.47 |
|
|
|
| chr3_+_22204475 | 1.45 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr19_+_43316957 | 1.43 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr10_-_38372418 | 1.43 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr2_+_7014241 | 1.42 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr4_-_20456437 | 1.40 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr5_-_36499541 | 1.36 |
|
|
|
| chr7_+_20651600 | 1.36 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr17_-_9903930 | 1.33 |
ENSDART00000166984
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr15_-_29229135 | 1.32 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr3_-_5523243 | 1.27 |
ENSDART00000159308
|
trim35-7
|
tripartite motif containing 35-7 |
| chr20_+_27088328 | 1.20 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr19_-_31455278 | 1.19 |
ENSDART00000133101
ENSDART00000136213 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr13_+_11696495 | 1.14 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr15_+_32391061 | 1.10 |
ENSDART00000152513
|
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr21_+_22088080 | 1.08 |
ENSDART00000130179
|
cul5b
|
cullin 5b |
| chr11_-_25223594 | 1.07 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr3_+_22204569 | 1.06 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr7_+_20651775 | 1.05 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr2_+_26002522 | 1.05 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr19_-_27811272 | 0.99 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr15_-_31296457 | 0.97 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr3_-_26060702 | 0.96 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr6_-_32424706 | 0.94 |
ENSDART00000078908
|
usp1
|
ubiquitin specific peptidase 1 |
| chr2_+_26002426 | 0.94 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr4_-_20456517 | 0.93 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr15_-_31296524 | 0.92 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr5_+_13370060 | 0.92 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr12_-_23244600 | 0.89 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr1_-_8801818 | 0.87 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr11_-_25224150 | 0.87 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr8_-_49356414 | 0.85 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr3_-_15517767 | 0.85 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr19_-_27811300 | 0.84 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr3_-_5523147 | 0.81 |
ENSDART00000159308
|
trim35-7
|
tripartite motif containing 35-7 |
| chr19_+_43316927 | 0.78 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr16_+_51290761 | 0.74 |
ENSDART00000058290
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr3_-_26060748 | 0.74 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr13_-_12534653 | 0.70 |
ENSDART00000079594
|
zgc:113201
|
zgc:113201 |
| chr16_+_39246663 | 0.68 |
ENSDART00000017017
|
zdhhc3a
|
zinc finger, DHHC-type containing 3a |
| chr12_-_18776533 | 0.66 |
ENSDART00000163582
|
desi1b
|
desumoylating isopeptidase 1b |
| chr18_-_44617805 | 0.64 |
ENSDART00000173095
ENSDART00000136420 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr3_-_36117170 | 0.59 |
ENSDART00000158701
ENSDART00000141106 |
gps1
|
G protein pathway suppressor 1 |
| chr5_+_13370237 | 0.58 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr6_-_32425011 | 0.53 |
ENSDART00000078908
|
usp1
|
ubiquitin specific peptidase 1 |
| chr10_-_2686054 | 0.52 |
ENSDART00000123754
|
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr1_+_5464124 | 0.51 |
ENSDART00000144559
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr21_-_13875945 | 0.37 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr1_+_58288476 | 0.30 |
|
|
|
| chr21_-_32747483 | 0.27 |
|
|
|
| chr18_-_34573536 | 0.26 |
ENSDART00000137101
|
ssr3
|
signal sequence receptor, gamma |
| chr8_-_43567869 | 0.26 |
ENSDART00000162770
|
ncor2
|
nuclear receptor corepressor 2 |
| chr24_-_32281707 | 0.23 |
ENSDART00000151845
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr6_+_11014565 | 0.22 |
ENSDART00000132677
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr23_+_36019942 | 0.20 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr17_-_37247455 | 0.20 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr8_+_47228598 | 0.12 |
ENSDART00000075068
|
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr7_+_6786457 | 0.12 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr11_-_25223723 | 0.08 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr4_-_39946152 | 0.06 |
|
|
|
| chr3_+_27188961 | 0.06 |
|
|
|
| chr19_-_37921386 | 0.06 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr4_-_5904787 | 0.02 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0003334 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.4 | 2.4 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 2.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.1 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.3 | 1.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.6 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 2.3 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 2.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 4.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 3.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 3.2 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.1 | 1.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 2.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 3.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 6.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 4.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 0.7 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 3.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.4 | 2.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.4 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 1.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 2.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 1.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 3.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 1.5 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.7 | GO:0045547 | polyprenyltransferase activity(GO:0002094) dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 4.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 0.7 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.1 | 4.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.2 | GO:0101005 | cysteine-type endopeptidase activity(GO:0004197) thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 6.9 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |