Project
DANIO-CODE
Navigation
Downloads
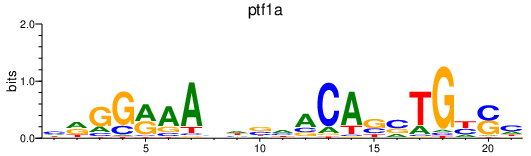
Results for ptf1a
Z-value: 0.71
Transcription factors associated with ptf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ptf1a
|
ENSDARG00000014479 | pancreas associated transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ptf1a | dr10_dc_chr2_+_29727235_29727242 | -0.69 | 2.8e-03 | Click! |
Activity profile of ptf1a motif
Sorted Z-values of ptf1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ptf1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_45182378 | 2.24 |
ENSDART00000161815
|
polm
|
polymerase (DNA directed), mu |
| chr22_+_22413803 | 1.62 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr22_-_20670164 | 1.58 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr25_+_33787063 | 1.49 |
|
|
|
| chr23_-_32267833 | 1.40 |
|
|
|
| chr3_+_17728616 | 1.37 |
ENSDART00000167953
|
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr4_+_48372465 | 1.26 |
|
|
|
| chr11_-_15886860 | 1.25 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr11_-_27715592 | 1.08 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr10_+_16914003 | 1.01 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr7_-_41532268 | 0.99 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr9_+_188749 | 0.96 |
ENSDART00000172946
|
rrp1
|
ribosomal RNA processing 1 |
| chr11_-_27580252 | 0.91 |
ENSDART00000121847
|
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr7_+_15479700 | 0.90 |
|
|
|
| chr23_+_39231080 | 0.82 |
|
|
|
| chr15_+_35076414 | 0.81 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr22_-_36721626 | 0.76 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr22_+_22413878 | 0.76 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr24_+_39631194 | 0.70 |
ENSDART00000013894
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr15_+_32053944 | 0.66 |
ENSDART00000175828
|
brca2
|
breast cancer 2, early onset |
| chr17_-_51729568 | 0.66 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr7_+_71658178 | 0.65 |
|
|
|
| chr15_-_46594821 | 0.64 |
ENSDART00000045110
|
chd
|
chordin |
| chr17_-_51729707 | 0.63 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr7_+_71658255 | 0.63 |
|
|
|
| chr5_-_47419494 | 0.63 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr17_+_21458177 | 0.62 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr19_-_44154954 | 0.61 |
ENSDART00000151309
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr5_-_32290272 | 0.58 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr18_-_18952992 | 0.56 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr7_+_41532360 | 0.55 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr9_+_188598 | 0.54 |
ENSDART00000162764
|
rrp1
|
ribosomal RNA processing 1 |
| chr5_-_47419466 | 0.53 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr3_+_32802124 | 0.53 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr24_+_39630741 | 0.53 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr11_-_28508152 | 0.52 |
|
|
|
| chr18_-_2954175 | 0.51 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr18_+_50921419 | 0.50 |
ENSDART00000058468
ENSDART00000008696 |
cttn
|
cortactin |
| chr8_-_1586037 | 0.49 |
ENSDART00000142703
|
CU694486.1
|
ENSDARG00000091948 |
| chr15_+_12504537 | 0.48 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr3_+_32802899 | 0.47 |
|
|
|
| chr19_-_44154669 | 0.46 |
ENSDART00000147328
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr15_+_2512287 | 0.45 |
ENSDART00000035939
ENSDART00000157845 |
panx1a
|
pannexin 1a |
| chr18_+_5260404 | 0.44 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr17_-_14443647 | 0.43 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8-associated membrane protein |
| chr3_+_37507573 | 0.43 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr7_+_15197911 | 0.43 |
ENSDART00000046542
|
igf1rb
|
insulin-like growth factor 1b receptor |
| chr22_-_26231695 | 0.43 |
ENSDART00000142821
|
ccdc130
|
coiled-coil domain containing 130 |
| chr7_+_28906011 | 0.42 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr5_-_47419401 | 0.42 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr8_-_18849529 | 0.42 |
ENSDART00000100491
|
sh3gl1b
|
SH3-domain GRB2-like 1b |
| chr5_-_32290231 | 0.42 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr7_+_15479520 | 0.41 |
|
|
|
| chr15_+_12504492 | 0.39 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr15_+_12504756 | 0.39 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr22_-_36721592 | 0.39 |
ENSDART00000130171
|
ncl
|
nucleolin |
| chr5_+_44538762 | 0.39 |
ENSDART00000172702
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_-_16893974 | 0.38 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr5_+_44208458 | 0.38 |
|
|
|
| chr13_+_24467302 | 0.38 |
ENSDART00000158949
|
AL831745.3
|
ENSDARG00000101436 |
| chr7_+_69212246 | 0.36 |
ENSDART00000159799
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| KN150589v1_-_6303 | 0.35 |
|
|
|
| chr25_+_33787029 | 0.34 |
|
|
|
| chr6_-_7094468 | 0.32 |
|
|
|
| chr7_+_69211965 | 0.32 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr11_-_27580123 | 0.31 |
ENSDART00000121847
|
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr22_-_26231606 | 0.30 |
ENSDART00000060888
|
ccdc130
|
coiled-coil domain containing 130 |
| chr19_-_12293162 | 0.30 |
ENSDART00000142077
|
znf706
|
zinc finger protein 706 |
| chr6_-_10677028 | 0.30 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr2_-_23331500 | 0.29 |
ENSDART00000146497
|
mllt1b
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1b |
| chr7_-_41531991 | 0.29 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr18_+_17548213 | 0.29 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr6_+_54350379 | 0.26 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr10_-_15090551 | 0.26 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr8_-_53553764 | 0.25 |
ENSDART00000157521
|
actr8
|
ARP8 actin related protein 8 homolog |
| chr5_-_47419195 | 0.25 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr7_+_69212144 | 0.24 |
ENSDART00000159799
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr15_+_1569864 | 0.23 |
ENSDART00000056765
|
smc4
|
structural maintenance of chromosomes 4 |
| chr3_+_61960035 | 0.22 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr12_-_3904030 | 0.21 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr5_-_47419154 | 0.21 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr24_+_16402587 | 0.21 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr15_+_46093261 | 0.20 |
|
|
|
| chr1_-_28146457 | 0.19 |
ENSDART00000110270
ENSDART00000170797 |
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr8_+_16722505 | 0.18 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr18_-_2954048 | 0.17 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr15_-_35075545 | 0.17 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr1_-_5387933 | 0.15 |
ENSDART00000138891
ENSDART00000176913 |
pard3ba
|
par-3 family cell polarity regulator beta a |
| chr16_+_35952050 | 0.15 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr5_+_37497105 | 0.13 |
|
|
|
| chr12_-_9094888 | 0.13 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr18_-_1745485 | 0.12 |
|
|
|
| chr15_-_2556119 | 0.11 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr5_-_47419233 | 0.11 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr3_-_36117170 | 0.11 |
ENSDART00000158701
ENSDART00000141106 |
gps1
|
G protein pathway suppressor 1 |
| chr15_-_1569267 | 0.10 |
ENSDART00000133943
|
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| KN150262v1_+_4307 | 0.10 |
|
|
|
| chr6_-_7094523 | 0.09 |
|
|
|
| chr14_+_49180737 | 0.08 |
|
|
|
| chr14_+_49180922 | 0.07 |
|
|
|
| chr13_+_8735 | 0.06 |
ENSDART00000168769
ENSDART00000165772 |
ENSDARG00000103415
|
ENSDARG00000103415 |
| chr18_-_19501985 | 0.06 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr16_-_25748643 | 0.06 |
ENSDART00000132136
|
derl1
|
derlin 1 |
| chr5_+_57073554 | 0.05 |
ENSDART00000166098
ENSDART00000005090 |
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr8_-_1201388 | 0.05 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr14_-_29498646 | 0.05 |
|
|
|
| chr8_+_36467811 | 0.05 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr3_+_61960120 | 0.05 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr10_+_45213266 | 0.05 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr21_-_2033162 | 0.05 |
|
|
|
| chr7_+_49391403 | 0.04 |
ENSDART00000131210
|
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr18_-_5260386 | 0.04 |
ENSDART00000027115
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr1_+_25773300 | 0.03 |
ENSDART00000011809
|
uso1
|
USO1 vesicle transport factor |
| chr23_+_1721822 | 0.03 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr7_+_15197937 | 0.02 |
ENSDART00000046542
|
igf1rb
|
insulin-like growth factor 1b receptor |
| chr18_-_5822255 | 0.01 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 2.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0008585 | female gonad development(GO:0008585) |
| 0.1 | 1.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.3 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.4 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 2.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |