Project
DANIO-CODE
Navigation
Downloads
Results for rarga
Z-value: 0.71
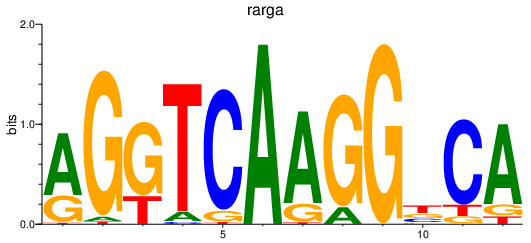
Transcription factors associated with rarga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarga
|
ENSDARG00000034117 | retinoic acid receptor gamma a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarga | dr10_dc_chr23_+_35748396_35748435 | 0.71 | 2.3e-03 | Click! |
Activity profile of rarga motif
Sorted Z-values of rarga motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rarga
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_49900124 | 2.77 |
ENSDART00000150204
|
atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr2_-_42384993 | 2.52 |
ENSDART00000141358
|
apom
|
apolipoprotein M |
| chr2_-_7693224 | 2.43 |
ENSDART00000167019
|
wu:fc46h12
|
wu:fc46h12 |
| chr25_+_4490129 | 1.83 |
|
|
|
| chr4_-_17420378 | 1.68 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr4_+_12033088 | 1.61 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr20_+_31366832 | 1.59 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr6_+_52235389 | 1.47 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr23_-_7283514 | 1.35 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr17_-_15490279 | 1.32 |
ENSDART00000156905
ENSDART00000161374 ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr6_-_18891908 | 1.24 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr4_-_21931540 | 1.22 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr8_+_48106836 | 1.22 |
ENSDART00000138717
|
si:ch211-263k4.2
|
si:ch211-263k4.2 |
| chr6_-_15526547 | 1.19 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr22_-_17570306 | 1.19 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr2_-_47578695 | 1.17 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr7_-_58427369 | 1.09 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr19_-_28382815 | 1.06 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr4_-_17640519 | 1.06 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr13_-_22731636 | 1.04 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr19_-_30816780 | 1.00 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr5_+_29578638 | 0.95 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr21_+_37757910 | 0.93 |
ENSDART00000162492
|
cox7b
|
cytochrome c oxidase subunit VIIb |
| chr24_-_14567403 | 0.91 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr11_+_36933891 | 0.91 |
ENSDART00000170542
|
ENSDARG00000100406
|
ENSDARG00000100406 |
| chr19_+_5156446 | 0.90 |
ENSDART00000151681
|
eno2
|
enolase 2 |
| chr22_-_18753903 | 0.85 |
ENSDART00000019235
|
atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr6_-_9964616 | 0.83 |
|
|
|
| chr2_+_33399405 | 0.83 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_26592356 | 0.79 |
ENSDART00000016608
|
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr2_+_24881103 | 0.78 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr3_-_18642744 | 0.77 |
ENSDART00000134208
|
hagh
|
hydroxyacylglutathione hydrolase |
| chr8_-_11806627 | 0.76 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr3_-_62156768 | 0.75 |
ENSDART00000140782
|
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr1_-_45941709 | 0.75 |
ENSDART00000053232
|
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr4_-_76620869 | 0.73 |
|
|
|
| chr24_+_9911748 | 0.71 |
|
|
|
| chr13_+_22134507 | 0.71 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr19_+_7255491 | 0.71 |
ENSDART00000151220
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr10_+_37983342 | 0.71 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr5_-_63487161 | 0.70 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr17_+_7438152 | 0.70 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr25_+_558191 | 0.67 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr14_-_1081316 | 0.67 |
|
|
|
| chr7_+_34235330 | 0.66 |
ENSDART00000173957
|
rfx7
|
regulatory factor X, 7 |
| chr19_+_30800672 | 0.65 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr14_-_5371581 | 0.65 |
ENSDART00000012116
|
tlx2
|
T-cell leukemia, homeobox 2 |
| chr24_+_19842162 | 0.64 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr23_-_19053587 | 0.61 |
|
|
|
| chr13_-_22713071 | 0.59 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr15_-_28687637 | 0.59 |
|
|
|
| chr5_-_34701639 | 0.58 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr5_-_54773058 | 0.58 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr20_+_51388214 | 0.58 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr20_+_26408687 | 0.57 |
|
|
|
| chr9_-_1969197 | 0.56 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr7_+_20092641 | 0.56 |
ENSDART00000139685
|
ponzr1
|
plac8 onzin related protein 1 |
| chr15_-_20997836 | 0.54 |
ENSDART00000152648
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr20_+_51388313 | 0.53 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr23_-_20026717 | 0.53 |
ENSDART00000153828
|
atp2b3b
|
ATPase, Ca++ transporting, plasma membrane 3b |
| chr20_+_51388660 | 0.52 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr14_+_29133320 | 0.51 |
ENSDART00000177215
|
ENSDARG00000037122
|
ENSDARG00000037122 |
| chr4_+_22754618 | 0.51 |
|
|
|
| chr19_-_22184035 | 0.50 |
|
|
|
| chr20_+_30894667 | 0.47 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr5_-_34701743 | 0.47 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr2_-_28154388 | 0.46 |
ENSDART00000161864
|
zgc:123035
|
zgc:123035 |
| chr3_-_30554400 | 0.46 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr24_-_16884946 | 0.46 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr6_+_22098053 | 0.46 |
ENSDART00000166987
|
rhot1b
|
ras homolog family member T1 |
| chr13_-_37320497 | 0.46 |
|
|
|
| chr13_+_15051177 | 0.45 |
ENSDART00000139308
|
pank2
|
pantothenate kinase 2 |
| chr2_-_10667156 | 0.45 |
ENSDART00000081196
|
scinlb
|
scinderin like b |
| chr2_-_52792076 | 0.45 |
ENSDART00000097716
|
zgc:136336
|
zgc:136336 |
| chr20_+_3260114 | 0.45 |
ENSDART00000067397
|
ndufaf7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr14_+_15728850 | 0.45 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr25_-_36713728 | 0.43 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr25_+_1747864 | 0.43 |
|
|
|
| chr10_-_43466392 | 0.42 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr24_+_3296536 | 0.42 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr14_-_33114284 | 0.41 |
ENSDART00000109615
|
tmem255a
|
transmembrane protein 255A |
| chr25_+_28329427 | 0.41 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr10_+_24476213 | 0.41 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr14_-_1081393 | 0.40 |
|
|
|
| chr14_+_45863932 | 0.40 |
ENSDART00000074216
|
higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr25_+_22474108 | 0.39 |
ENSDART00000161559
|
stra6
|
stimulated by retinoic acid 6 |
| chr23_+_35988395 | 0.39 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr14_-_2278018 | 0.39 |
ENSDART00000125674
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr13_-_508202 | 0.39 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr11_-_42933969 | 0.39 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr25_+_5567388 | 0.39 |
|
|
|
| chr20_+_51388752 | 0.39 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr19_+_23414927 | 0.38 |
ENSDART00000151090
|
gdf6b
|
growth differentiation factor 6b |
| chr3_-_30554490 | 0.37 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr23_+_24862663 | 0.37 |
ENSDART00000088697
|
olfml3a
|
olfactomedin-like 3a |
| chr10_-_4979688 | 0.36 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr5_-_55263153 | 0.36 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr5_-_64201886 | 0.36 |
ENSDART00000145819
|
krt1-c5
|
keratin, type 1, gene c5 |
| chr10_-_43466478 | 0.35 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr15_-_25592228 | 0.35 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr22_-_22696091 | 0.34 |
|
|
|
| chr3_+_26015378 | 0.34 |
|
|
|
| chr17_+_31604804 | 0.34 |
ENSDART00000111629
|
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr20_+_7337738 | 0.33 |
ENSDART00000165596
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr23_+_42370612 | 0.33 |
ENSDART00000161812
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr7_+_15781933 | 0.33 |
ENSDART00000161669
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr24_-_20513975 | 0.31 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr4_-_22950321 | 0.31 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr18_+_41243277 | 0.31 |
|
|
|
| chr10_-_16069787 | 0.30 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr21_-_11564398 | 0.30 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr4_-_13568659 | 0.29 |
ENSDART00000169582
|
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr13_-_23838192 | 0.29 |
|
|
|
| chr8_-_38316544 | 0.29 |
|
|
|
| chr14_+_5852428 | 0.29 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr9_-_35062828 | 0.29 |
ENSDART00000026378
|
slc25a6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr17_-_15490230 | 0.29 |
ENSDART00000156905
ENSDART00000161374 ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr11_+_30035395 | 0.28 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr3_+_21059221 | 0.27 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr3_-_36465046 | 0.27 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr20_+_54247734 | 0.27 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr11_+_22213887 | 0.26 |
ENSDART00000174683
|
ppfia4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr15_+_46206534 | 0.26 |
ENSDART00000099202
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr25_+_16796766 | 0.25 |
|
|
|
| chr11_+_37453275 | 0.25 |
ENSDART00000050296
|
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr10_+_20112765 | 0.25 |
ENSDART00000027612
|
xpo7
|
exportin 7 |
| chr7_+_26278650 | 0.25 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr19_+_30800724 | 0.24 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr16_-_22947908 | 0.24 |
ENSDART00000133910
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr8_-_15955746 | 0.24 |
ENSDART00000132908
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr3_-_25238896 | 0.24 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr18_-_22764560 | 0.23 |
ENSDART00000009912
|
hsf4
|
heat shock transcription factor 4 |
| chr22_-_17570275 | 0.22 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr2_-_6351592 | 0.22 |
|
|
|
| chr21_+_19889712 | 0.22 |
ENSDART00000147294
|
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr19_+_41630199 | 0.22 |
|
|
|
| chr14_+_30455165 | 0.22 |
ENSDART00000144817
|
cfl1
|
cofilin 1 |
| chr12_-_18286666 | 0.22 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr23_-_19577146 | 0.21 |
ENSDART00000143288
|
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr23_+_37807239 | 0.21 |
ENSDART00000129531
|
CR381544.1
|
ENSDARG00000088187 |
| chr19_+_43524904 | 0.21 |
|
|
|
| chr8_+_24720852 | 0.21 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr25_+_1511300 | 0.21 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr20_+_26196386 | 0.21 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr17_-_43296680 | 0.21 |
ENSDART00000176637
|
EML5
|
echinoderm microtubule associated protein like 5 |
| chr7_-_15781823 | 0.21 |
ENSDART00000002498
|
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr9_+_31469848 | 0.21 |
|
|
|
| chr16_-_7310925 | 0.20 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr8_-_50299273 | 0.20 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr20_-_37673852 | 0.20 |
|
|
|
| chr6_-_10855968 | 0.19 |
ENSDART00000127209
|
pttg1ipa
|
PTTG1 interacting protein a |
| chr11_-_42934175 | 0.18 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr22_-_17571993 | 0.18 |
|
|
|
| chr9_+_21524891 | 0.17 |
|
|
|
| chr8_-_38316609 | 0.17 |
|
|
|
| chr7_-_51210658 | 0.17 |
ENSDART00000174026
|
nhsl2
|
NHS-like 2 |
| chr3_-_62156409 | 0.17 |
ENSDART00000101870
ENSDART00000175992 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr14_-_33114322 | 0.16 |
ENSDART00000173267
|
tmem255a
|
transmembrane protein 255A |
| chr2_-_24330667 | 0.16 |
|
|
|
| chr5_+_69096325 | 0.15 |
ENSDART00000014649
|
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr19_+_14197020 | 0.14 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr16_-_2269074 | 0.13 |
|
|
|
| chr3_-_25239180 | 0.12 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr12_+_3875774 | 0.12 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr17_+_51135365 | 0.12 |
ENSDART00000025229
|
adi1
|
acireductone dioxygenase 1 |
| chr13_+_33173790 | 0.12 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr15_+_22078226 | 0.11 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr23_-_31721545 | 0.11 |
ENSDART00000133569
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_-_34701683 | 0.11 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr4_-_13568618 | 0.10 |
ENSDART00000169582
|
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr14_-_35073975 | 0.10 |
ENSDART00000145033
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr13_+_17337382 | 0.10 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr14_-_24814149 | 0.09 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| KN150170v1_+_44642 | 0.09 |
ENSDART00000177674
|
CABZ01092074.3
|
ENSDARG00000109197 |
| chr4_+_6726443 | 0.09 |
ENSDART00000150389
|
tmem168a
|
transmembrane protein 168a |
| chr2_-_24633983 | 0.09 |
ENSDART00000128784
ENSDART00000141922 ENSDART00000146213 |
vmhcl
|
ventricular myosin heavy chain-like |
| chr13_+_33173742 | 0.08 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr17_+_7437994 | 0.08 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr5_-_52357382 | 0.08 |
ENSDART00000157978
|
mrps30
|
mitochondrial ribosomal protein S30 |
| chr7_+_20595544 | 0.08 |
ENSDART00000173495
|
gigyf1a
|
GRB10 interacting GYF protein 1a |
| chr9_-_27939140 | 0.07 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr20_+_36779159 | 0.06 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr21_-_11564441 | 0.05 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr7_-_31346991 | 0.05 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr23_+_35988426 | 0.04 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr2_-_24330786 | 0.04 |
|
|
|
| chr17_-_9774006 | 0.03 |
ENSDART00000149640
|
egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr25_-_3619663 | 0.03 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr14_-_49044809 | 0.03 |
ENSDART00000169300
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr13_-_15863571 | 0.02 |
ENSDART00000016430
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr13_+_40208832 | 0.02 |
ENSDART00000074950
|
slc25a28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr6_-_10600358 | 0.02 |
ENSDART00000135065
|
atp5g3b
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c3 (subunit 9) b |
| chr6_-_10676890 | 0.01 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr5_+_62696901 | 0.01 |
ENSDART00000124616
|
rab14
|
RAB14, member RAS oncogene family |
| chr2_+_26824005 | 0.01 |
ENSDART00000172669
ENSDART00000056795 ENSDART00000144837 |
hectd3
|
HECT domain containing 3 |
| chr14_-_35074225 | 0.01 |
ENSDART00000129460
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr7_-_41871376 | 0.00 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr17_+_7438085 | 0.00 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0071604 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.3 | 1.7 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.3 | 1.2 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.2 | 0.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.8 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.7 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.1 | 0.7 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.6 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.2 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 1.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.3 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 1.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 2.4 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.1 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.8 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.4 | GO:1901880 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) actin filament capping(GO:0051693) regulation of protein depolymerization(GO:1901879) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.7 | GO:0033339 | pectoral fin development(GO:0033339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 9.4 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 1.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.6 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.2 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.7 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 2.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.2 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 3.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |