Project
DANIO-CODE
Navigation
Downloads
Results for rargb_pparab
Z-value: 1.60
Transcription factors associated with rargb_pparab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
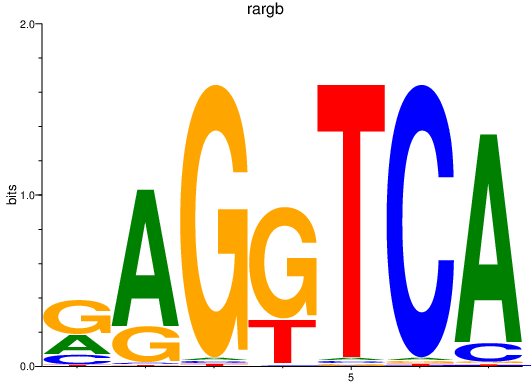
rargb
|
ENSDARG00000054003 | retinoic acid receptor, gamma b |
|
pparab
|
ENSDARG00000054323 | peroxisome proliferator-activated receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pparab | dr10_dc_chr25_+_2107536_2107580 | 0.62 | 1.0e-02 | Click! |
Activity profile of rargb_pparab motif
Sorted Z-values of rargb_pparab motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rargb_pparab
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_44817648 | 4.41 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr11_+_11217547 | 4.38 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr5_+_40722565 | 4.28 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr3_+_33168814 | 4.24 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr13_-_39033893 | 4.18 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr22_-_17570306 | 4.10 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr18_+_62936 | 4.02 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr2_+_42342148 | 3.91 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr22_+_33177432 | 3.89 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr13_-_37001997 | 3.88 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_+_29716279 | 3.84 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr2_-_47578695 | 3.81 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr12_-_5085227 | 3.76 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr3_-_48865474 | 3.70 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_-_39491932 | 3.70 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr15_-_29454583 | 3.68 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr20_+_2255204 | 3.65 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr13_+_1595986 | 3.63 |
|
|
|
| chr3_+_39425125 | 3.59 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr3_+_23587156 | 3.57 |
|
|
|
| chr12_+_27032862 | 3.52 |
|
|
|
| chr10_-_44180588 | 3.49 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr17_+_16067769 | 3.41 |
ENSDART00000155292
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr7_+_25758469 | 3.23 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr24_-_12794057 | 3.16 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr4_-_16461748 | 3.16 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr9_+_42112576 | 3.16 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr13_-_37144297 | 3.15 |
|
|
|
| chr17_+_8874210 | 3.08 |
|
|
|
| chr5_-_46382728 | 3.07 |
|
|
|
| chr14_-_48951428 | 3.03 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr25_+_4490129 | 3.01 |
|
|
|
| chr2_-_43110857 | 2.98 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr15_+_24708696 | 2.96 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr4_-_1346961 | 2.85 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr3_-_18426055 | 2.84 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr19_-_7069920 | 2.80 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr6_-_13654186 | 2.77 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr18_+_45893199 | 2.76 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr6_+_3549861 | 2.69 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr2_+_45228446 | 2.69 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr14_+_14272033 | 2.65 |
ENSDART00000167483
|
plp1a
|
proteolipid protein 1a |
| chr9_-_41982635 | 2.64 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr24_-_21778717 | 2.64 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr6_-_39315024 | 2.63 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr7_+_65532390 | 2.61 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr10_+_6316944 | 2.52 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr23_-_9924987 | 2.51 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr18_+_9413588 | 2.49 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr20_-_45819128 | 2.47 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| KN150207v1_-_1208 | 2.44 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr7_+_40604000 | 2.40 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr3_-_45779817 | 2.39 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr7_+_25641990 | 2.38 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr15_-_25582891 | 2.38 |
|
|
|
| chr7_+_40603904 | 2.37 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr4_-_76620869 | 2.31 |
|
|
|
| chr12_-_457997 | 2.30 |
ENSDART00000143232
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr10_-_44441481 | 2.29 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr22_+_13952935 | 2.29 |
ENSDART00000080313
|
arl4ca
|
ADP-ribosylation factor-like 4Ca |
| chr2_+_38057483 | 2.26 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr2_-_24357532 | 2.24 |
ENSDART00000137065
|
col15a1b
|
collagen, type XV, alpha 1b |
| chr23_+_36079164 | 2.23 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr12_+_28739504 | 2.21 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr21_-_22510751 | 2.21 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr11_+_30482530 | 2.20 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr17_+_381715 | 2.20 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr1_+_16681778 | 2.19 |
|
|
|
| chr5_-_54773058 | 2.18 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr20_-_53162473 | 2.17 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr23_-_21520413 | 2.17 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr1_+_23093114 | 2.16 |
|
|
|
| chr5_+_26840147 | 2.14 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr25_-_13518274 | 2.14 |
ENSDART00000165510
|
fa2h
|
fatty acid 2-hydroxylase |
| chr2_+_52473102 | 2.13 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr15_-_23711689 | 2.13 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr2_-_24275851 | 2.12 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr25_-_33920256 | 2.11 |
|
|
|
| chr22_+_10172134 | 2.09 |
ENSDART00000132641
|
pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr13_+_22350043 | 2.09 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr9_+_9524547 | 2.08 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr6_-_49900124 | 2.07 |
ENSDART00000150204
|
atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr7_+_19765237 | 2.07 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr22_-_2870591 | 2.05 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr7_+_48532749 | 2.00 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr16_-_39181608 | 2.00 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr6_-_40724581 | 1.99 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr13_-_219661 | 1.99 |
ENSDART00000133731
|
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr8_-_23102555 | 1.98 |
|
|
|
| chr9_-_22528568 | 1.98 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr9_-_48152388 | 1.98 |
|
|
|
| chr19_-_47994946 | 1.96 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr13_+_30546398 | 1.95 |
|
|
|
| chr17_+_135165 | 1.94 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr25_-_18855316 | 1.93 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr17_-_31041977 | 1.93 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr9_-_53220424 | 1.90 |
|
|
|
| chr5_-_21520111 | 1.90 |
|
|
|
| chr19_-_5441932 | 1.88 |
ENSDART00000105034
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr25_+_35901081 | 1.88 |
ENSDART00000042271
ENSDART00000158027 |
irx3b
|
iroquois homeobox 3b |
| chr1_-_40208544 | 1.87 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr22_+_16511562 | 1.86 |
|
|
|
| chr22_+_26543146 | 1.85 |
|
|
|
| KN149795v1_-_24393 | 1.85 |
|
|
|
| chr16_-_17439735 | 1.83 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr11_-_6960107 | 1.80 |
ENSDART00000171255
|
COMP
|
cartilage oligomeric matrix protein |
| chr19_-_7069850 | 1.80 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr12_+_30119269 | 1.79 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr23_-_35657313 | 1.79 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr21_+_5483017 | 1.78 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr12_-_29190576 | 1.77 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr17_+_443558 | 1.75 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr1_-_40208469 | 1.74 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr2_+_393439 | 1.74 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr16_-_45943282 | 1.74 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr20_-_29580624 | 1.74 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr13_-_39821399 | 1.74 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr21_-_3301751 | 1.73 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr19_-_39048324 | 1.72 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr22_-_26971466 | 1.72 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr13_-_2259405 | 1.71 |
ENSDART00000166456
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr4_-_25847471 | 1.71 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr14_+_38375351 | 1.71 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr23_+_26215871 | 1.71 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr2_-_14757989 | 1.71 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr10_-_43711606 | 1.70 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr19_-_5416317 | 1.70 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr10_+_4717800 | 1.70 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr5_+_1223598 | 1.70 |
ENSDART00000159783
ENSDART00000026535 ENSDART00000168130 ENSDART00000147972 |
dnm1a
|
dynamin 1a |
| chr17_+_12254292 | 1.70 |
ENSDART00000160722
|
khk
|
ketohexokinase |
| chr1_+_25157675 | 1.69 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr4_-_935448 | 1.68 |
ENSDART00000171855
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr23_+_6298911 | 1.66 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr24_-_16884946 | 1.66 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr17_+_134921 | 1.65 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr6_-_37481918 | 1.65 |
ENSDART00000130359
|
si:dkey-66a8.7
|
si:dkey-66a8.7 |
| chr5_+_26832823 | 1.64 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr8_+_48488009 | 1.64 |
|
|
|
| chr1_-_18709814 | 1.62 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr13_-_37002066 | 1.61 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr24_+_16249188 | 1.60 |
ENSDART00000164516
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr11_+_43559400 | 1.59 |
|
|
|
| chr7_+_5372830 | 1.59 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr11_-_7310284 | 1.59 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr7_-_52434786 | 1.58 |
ENSDART00000131282
|
tcf12
|
transcription factor 12 |
| chr12_+_25006393 | 1.57 |
ENSDART00000050070
|
pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr9_-_21257082 | 1.56 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr7_+_25642061 | 1.56 |
ENSDART00000135728
|
hmgb3a
|
high mobility group box 3a |
| chr9_-_1969197 | 1.55 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr23_+_4083697 | 1.54 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr3_-_32795866 | 1.53 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr21_+_15613081 | 1.53 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr20_-_39200252 | 1.53 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr2_-_30005139 | 1.52 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr17_-_17744885 | 1.52 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr19_+_41274734 | 1.52 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr10_+_34372205 | 1.51 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr19_+_3925188 | 1.51 |
|
|
|
| chr9_-_34491458 | 1.50 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr3_+_47735048 | 1.49 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr19_+_4996328 | 1.49 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr14_-_46359248 | 1.49 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr18_-_15963983 | 1.48 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr8_-_14146621 | 1.47 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr11_-_33925792 | 1.47 |
ENSDART00000172978
|
atp13a3
|
ATPase type 13A3 |
| chr23_+_18796386 | 1.46 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr18_+_36650892 | 1.46 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr10_-_39210658 | 1.46 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr13_+_1377875 | 1.46 |
|
|
|
| chr17_-_38831108 | 1.45 |
ENSDART00000124041
|
c17h14orf159
|
c17h14orf159 homolog (H. sapiens) |
| chr4_+_12033088 | 1.45 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr5_-_63031745 | 1.45 |
|
|
|
| chr9_+_9524688 | 1.45 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr1_-_46233556 | 1.45 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr18_-_48498261 | 1.44 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr14_+_35906366 | 1.44 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr11_+_7148830 | 1.44 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr3_-_26393945 | 1.44 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr7_+_25861386 | 1.44 |
ENSDART00000129834
|
nat16
|
N-acetyltransferase 16 |
| chr9_-_711269 | 1.43 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr16_+_25491981 | 1.43 |
ENSDART00000086333
|
jarid2a
|
jumonji, AT rich interactive domain 2a |
| chr25_-_30818836 | 1.43 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr25_+_21732255 | 1.41 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr14_+_34174018 | 1.41 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr5_+_26925238 | 1.41 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr7_-_52848084 | 1.41 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr22_+_18218979 | 1.41 |
|
|
|
| chr7_+_26980284 | 1.40 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr8_+_49582361 | 1.40 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr22_+_30234718 | 1.40 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr16_-_10404109 | 1.40 |
|
|
|
| chr15_-_35563367 | 1.39 |
|
|
|
| chr16_-_7321943 | 1.39 |
ENSDART00000149260
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr17_-_5426195 | 1.38 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr12_+_42244935 | 1.38 |
ENSDART00000166484
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr2_-_20941256 | 1.37 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr21_-_2147085 | 1.36 |
ENSDART00000169262
|
zgc:163077
|
zgc:163077 |
| chr17_+_30431520 | 1.36 |
ENSDART00000153826
|
lpin1
|
lipin 1 |
| chr8_+_25714744 | 1.36 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr14_-_46359062 | 1.35 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 1.5 | 4.4 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 1.0 | 3.1 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.9 | 2.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.9 | 2.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.8 | 3.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.8 | 3.2 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.8 | 2.3 | GO:0061130 | branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.8 | 4.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 2.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.7 | 2.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.6 | 1.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.6 | 4.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.6 | 1.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.5 | 5.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.5 | 2.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.5 | 1.5 | GO:0032675 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) |
| 0.5 | 1.4 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.5 | 1.9 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.5 | 1.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.5 | 3.8 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.5 | 3.6 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.4 | 3.5 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.3 | GO:0086010 | mechanosensory behavior(GO:0007638) membrane depolarization during action potential(GO:0086010) |
| 0.4 | 3.8 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.4 | 1.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 1.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 1.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.4 | 0.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 2.8 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 1.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.5 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.4 | 2.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.4 | 2.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 1.1 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 1.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.3 | 1.7 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.3 | 1.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 1.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 0.9 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.3 | 2.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 2.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.3 | 2.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 1.4 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.3 | 1.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 0.5 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.3 | 1.6 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.3 | 1.1 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.3 | 1.0 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.2 | 0.7 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 2.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 0.7 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 1.3 | GO:0009110 | vitamin biosynthetic process(GO:0009110) |
| 0.2 | 2.9 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 1.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 1.0 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 1.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 3.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.1 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.9 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 2.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 0.5 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.7 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) negative regulation of cell aging(GO:0090344) regulation of cellular senescence(GO:2000772) |
| 0.2 | 0.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 0.7 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 2.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 0.5 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.2 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 10.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.2 | 0.5 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.4 | GO:0030852 | regulation of collagen metabolic process(GO:0010712) regulation of granulocyte differentiation(GO:0030852) plasminogen activation(GO:0031639) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 3.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 1.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.6 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 2.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.9 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.6 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 0.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.0 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 1.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 1.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 9.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 1.8 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 1.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0007183 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 3.1 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.1 | 1.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 6.5 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 2.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.2 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:1902946 | protein localization to endosome(GO:0036010) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 2.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 8.6 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 1.0 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 3.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.3 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:1900076 | regulation of insulin receptor signaling pathway(GO:0046626) regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.1 | 0.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.6 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 2.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.6 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 1.3 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 1.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.2 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.3 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.4 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 1.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 2.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.7 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 2.9 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 2.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 0.8 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.1 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.4 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 2.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.1 | GO:0046460 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.1 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.3 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 7.5 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 1.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 2.2 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.7 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 1.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.8 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 1.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 3.3 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 0.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.8 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.1 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.9 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 1.9 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0070476 | peptidyl-glutamine methylation(GO:0018364) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0010878 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 2.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.9 | 2.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.5 | 5.5 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 2.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 3.9 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.4 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 1.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 2.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 3.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.3 | 4.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 10.5 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.3 | 2.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 9.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 10.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 6.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 0.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 4.1 | GO:0044420 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.2 | 3.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 6.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 5.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 3.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 36.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 16.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 3.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.4 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0045177 | apical plasma membrane(GO:0016324) apical part of cell(GO:0045177) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 2.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.8 | 4.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.8 | 3.2 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 3.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.7 | 4.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.7 | 2.7 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.6 | 1.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 2.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.5 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 2.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.5 | 1.4 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.5 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 4.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.5 | 3.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 3.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 6.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 0.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.4 | 1.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.4 | 2.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 2.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 8.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 4.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.3 | 1.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 1.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 3.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 2.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 0.8 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.3 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 0.8 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.2 | 1.0 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.2 | 3.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 1.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 3.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 4.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 1.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.6 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.7 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.2 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 2.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 8.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.7 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 2.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.4 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 1.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.3 | GO:0070699 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 2.8 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 14.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 6.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 1.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 6.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0030151 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 9.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 22.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 4.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 4.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 3.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 3.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 2.7 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.3 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.8 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.2 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 9.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |