Project
DANIO-CODE
Navigation
Downloads
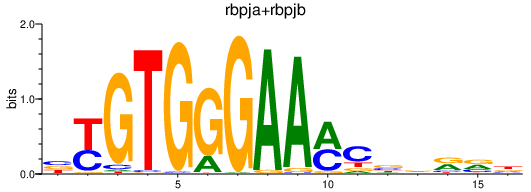
Results for rbpja+rbpjb
Z-value: 0.81
Transcription factors associated with rbpja+rbpjb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpja
|
ENSDARG00000003398 | recombination signal binding protein for immunoglobulin kappa J region a |
|
rbpjb
|
ENSDARG00000052091 | recombination signal binding protein for immunoglobulin kappa J region b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpja | dr10_dc_chr1_+_13768098_13768238 | 0.82 | 8.6e-05 | Click! |
| rbpjb | dr10_dc_chr7_+_61770242_61770424 | -0.56 | 2.5e-02 | Click! |
Activity profile of rbpja+rbpjb motif
Sorted Z-values of rbpja+rbpjb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpja+rbpjb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_2000017 | 3.50 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr19_+_32000438 | 3.43 |
ENSDART00000150910
|
gmnn
|
geminin, DNA replication inhibitor |
| chr20_+_14893184 | 3.26 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr17_-_2396632 | 2.39 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr6_-_53145464 | 1.85 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr16_+_9823111 | 1.68 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr12_-_28680035 | 1.65 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr17_+_16038103 | 1.58 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_-_31738565 | 1.58 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_-_9212139 | 1.56 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr5_-_35997198 | 1.42 |
ENSDART00000031270
|
rhogc
|
ras homolog gene family, member Gc |
| chr15_-_31266460 | 1.39 |
ENSDART00000157145
ENSDART00000155473 |
ksr1b
|
kinase suppressor of ras 1b |
| chr5_-_35997345 | 1.37 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr12_-_46943717 | 1.35 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr6_-_2000167 | 1.29 |
ENSDART00000160475
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr8_+_36522231 | 1.26 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr6_-_9459739 | 1.25 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr3_-_3646681 | 1.13 |
ENSDART00000165648
ENSDART00000046454 ENSDART00000115282 ENSDART00000164463 |
ENSDARG00000076771
|
ENSDARG00000076771 |
| chr11_-_3516034 | 1.11 |
ENSDART00000009788
|
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr8_-_22266041 | 1.11 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr14_+_41039244 | 1.07 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr13_-_6123969 | 1.07 |
ENSDART00000161163
|
tuba4l
|
tubulin, alpha 4 like |
| chr25_+_20174490 | 1.07 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr5_-_35997304 | 1.05 |
ENSDART00000031270
|
rhogc
|
ras homolog gene family, member Gc |
| chr23_+_25808393 | 1.04 |
|
|
|
| chr9_+_38672040 | 1.03 |
ENSDART00000157556
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr11_+_41777220 | 1.03 |
ENSDART00000172846
|
camta1
|
calmodulin binding transcription activator 1 |
| chr8_-_22265875 | 1.02 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr2_+_8314513 | 1.01 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr6_+_3556296 | 1.01 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr21_-_9633519 | 1.01 |
|
|
|
| chr6_-_53145759 | 1.00 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| KN150455v1_-_14200 | 0.99 |
|
|
|
| chr21_+_39649559 | 0.97 |
ENSDART00000100240
|
rnmtl1b
|
RNA methyltransferase like 1b |
| chr19_+_14132374 | 0.95 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr25_+_30627892 | 0.95 |
|
|
|
| chr19_-_12728868 | 0.94 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr19_+_14132473 | 0.91 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| KN150455v1_-_14097 | 0.90 |
|
|
|
| chr5_-_17372745 | 0.90 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr10_-_10907273 | 0.90 |
|
|
|
| chr25_+_30627755 | 0.88 |
|
|
|
| chr5_+_4092360 | 0.86 |
ENSDART00000158826
|
CABZ01058650.1
|
ENSDARG00000101063 |
| chr24_+_39339391 | 0.85 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr17_+_44642582 | 0.81 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr5_+_40543392 | 0.81 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr13_-_37339532 | 0.78 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr6_-_53145582 | 0.78 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr12_-_37274632 | 0.77 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr12_+_37039816 | 0.77 |
ENSDART00000162922
|
CR456627.1
|
ENSDARG00000104543 |
| chr24_+_7855521 | 0.75 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr5_+_6005049 | 0.74 |
|
|
|
| chr17_+_45489354 | 0.74 |
ENSDART00000103493
|
ENSDARG00000070487
|
ENSDARG00000070487 |
| chr6_+_58528496 | 0.74 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr3_-_86374 | 0.73 |
ENSDART00000138302
|
zgc:110249
|
zgc:110249 |
| chr16_-_25765512 | 0.73 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr16_-_25765396 | 0.73 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr24_+_24025923 | 0.72 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr1_-_30301568 | 0.71 |
ENSDART00000132466
|
si:ch211-269i23.2
|
si:ch211-269i23.2 |
| chr2_-_44123230 | 0.71 |
|
|
|
| chr16_-_32865452 | 0.71 |
|
|
|
| chr10_-_14985202 | 0.70 |
ENSDART00000143608
|
smad2
|
SMAD family member 2 |
| chr24_-_20946041 | 0.70 |
ENSDART00000140786
|
qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr16_-_25765322 | 0.69 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr20_-_9212089 | 0.68 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr5_+_37497397 | 0.68 |
|
|
|
| chr6_-_30696540 | 0.67 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr20_+_14893374 | 0.67 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr22_+_1451958 | 0.65 |
|
|
|
| chr13_-_37339582 | 0.65 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr11_-_27290434 | 0.64 |
ENSDART00000173444
|
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr8_+_19945528 | 0.64 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr2_+_8314372 | 0.63 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr24_+_20514547 | 0.61 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr8_+_13757024 | 0.61 |
|
|
|
| chr11_-_3593758 | 0.61 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr5_+_60677040 | 0.61 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr10_-_10907343 | 0.60 |
|
|
|
| chr5_+_40543353 | 0.60 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr16_-_25765583 | 0.59 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr11_-_1382493 | 0.58 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr17_+_4902984 | 0.58 |
ENSDART00000064313
ENSDART00000121806 |
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr19_+_7082199 | 0.57 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr16_-_42203272 | 0.57 |
|
|
|
| chr21_-_32048768 | 0.57 |
ENSDART00000146079
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr12_+_33359784 | 0.57 |
ENSDART00000007053
|
narf
|
nuclear prelamin A recognition factor |
| chr6_+_49411169 | 0.56 |
|
|
|
| chr24_-_31203020 | 0.55 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr3_+_62116865 | 0.55 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr17_-_6351209 | 0.55 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr15_-_14100192 | 0.55 |
|
|
|
| chr2_+_44928188 | 0.55 |
ENSDART00000147292
|
ece2a
|
endothelin converting enzyme 2a |
| chr20_+_23602537 | 0.54 |
|
|
|
| chr15_-_25499679 | 0.53 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr12_-_28680442 | 0.53 |
ENSDART00000178777
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr5_-_23171454 | 0.51 |
ENSDART00000135153
|
TBC1D8B
|
TBC1 domain family member 8B |
| chr7_-_41446321 | 0.51 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr16_-_25765542 | 0.51 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr11_-_1382526 | 0.51 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr2_+_8314844 | 0.50 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr9_-_30692223 | 0.49 |
|
|
|
| chr15_+_44168674 | 0.49 |
|
|
|
| chr7_+_17460777 | 0.49 |
|
|
|
| chr23_-_465425 | 0.48 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr17_-_6351014 | 0.48 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr11_-_3593791 | 0.48 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr20_-_54285455 | 0.48 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr24_+_8596265 | 0.47 |
ENSDART00000113493
|
tmem14ca
|
transmembrane protein 14Ca |
| chr5_+_1461715 | 0.46 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr3_+_36504142 | 0.46 |
ENSDART00000170013
|
gspt1l
|
G1 to S phase transition 1, like |
| chr8_-_22265802 | 0.44 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr17_+_4903106 | 0.44 |
ENSDART00000064313
ENSDART00000121806 |
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr18_-_17085956 | 0.44 |
ENSDART00000042496
|
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr24_+_39139519 | 0.44 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr24_+_24025701 | 0.42 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr23_-_465261 | 0.41 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr17_+_5454020 | 0.41 |
ENSDART00000097455
|
fam167aa
|
family with sequence similarity 167, member Aa |
| chr24_-_31203112 | 0.41 |
ENSDART00000167837
|
tmem56a
|
transmembrane protein 56a |
| chr17_-_31702025 | 0.41 |
ENSDART00000136199
|
dtd2
|
D-tyrosyl-tRNA deacylase 2 |
| chr20_+_53563189 | 0.41 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr23_+_17912784 | 0.40 |
ENSDART00000104647
|
prim1
|
primase, DNA, polypeptide 1 |
| chr8_+_15240122 | 0.39 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr20_+_5022501 | 0.39 |
ENSDART00000162678
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr15_-_14100434 | 0.39 |
|
|
|
| chr2_-_55584106 | 0.38 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr7_-_28300008 | 0.37 |
|
|
|
| chr21_+_33999164 | 0.37 |
ENSDART00000178338
|
map4k6
|
mitogen-activated protein kinase kinase kinase kinase 6 |
| chr15_+_1238560 | 0.36 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr16_+_8225781 | 0.36 |
ENSDART00000139432
|
pomgnt2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr16_-_25765623 | 0.36 |
ENSDART00000015302
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr13_-_2812369 | 0.36 |
|
|
|
| chr4_-_71920187 | 0.34 |
ENSDART00000157532
|
CU570689.2
|
ENSDARG00000105713 |
| chr12_+_25132724 | 0.34 |
ENSDART00000127454
ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr4_+_73504617 | 0.34 |
ENSDART00000148575
|
nup50
|
nucleoporin 50 |
| KN150311v1_-_13973 | 0.33 |
ENSDART00000160072
|
snx1b
|
sorting nexin 1b |
| chr25_+_2172612 | 0.33 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr19_+_42863138 | 0.32 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr9_-_28444210 | 0.31 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr3_-_52837780 | 0.31 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr9_-_25555518 | 0.31 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr17_+_44642465 | 0.31 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr12_+_40536264 | 0.30 |
|
|
|
| chr5_+_19815701 | 0.30 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr3_+_10670764 | 0.30 |
ENSDART00000129257
|
tmem220
|
transmembrane protein 220 |
| chr25_+_34410718 | 0.29 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr5_+_37497105 | 0.29 |
|
|
|
| chr5_+_44208406 | 0.29 |
|
|
|
| chr15_-_14147062 | 0.29 |
ENSDART00000147796
|
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr4_-_2742799 | 0.28 |
|
|
|
| chr8_-_36522230 | 0.28 |
ENSDART00000132804
|
ccdc157
|
coiled-coil domain containing 157 |
| chr11_+_15346076 | 0.28 |
ENSDART00000155655
|
cdk5rap1
|
CDK5 regulatory subunit associated protein 1 |
| chr3_-_30927275 | 0.27 |
ENSDART00000112266
|
armc5
|
armadillo repeat containing 5 |
| chr1_-_51086075 | 0.27 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr4_-_2742712 | 0.27 |
|
|
|
| chr7_-_73622399 | 0.26 |
ENSDART00000126541
|
zgc:173552
|
zgc:173552 |
| chr3_-_96489 | 0.26 |
|
|
|
| chr19_+_40661661 | 0.26 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr9_-_7110860 | 0.25 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr10_+_573116 | 0.24 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr21_+_33999222 | 0.23 |
ENSDART00000178338
|
map4k6
|
mitogen-activated protein kinase kinase kinase kinase 6 |
| chr2_-_8314813 | 0.22 |
|
|
|
| chr12_-_37274547 | 0.22 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| KN150455v1_-_13937 | 0.22 |
|
|
|
| chr1_-_30301438 | 0.21 |
ENSDART00000132466
|
si:ch211-269i23.2
|
si:ch211-269i23.2 |
| chr23_-_14460492 | 0.20 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr12_+_25132623 | 0.20 |
ENSDART00000127454
ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr14_+_41039161 | 0.19 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr3_+_33990660 | 0.19 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr19_+_41935890 | 0.18 |
ENSDART00000115123
|
crtc2
|
CREB regulated transcription coactivator 2 |
| chr23_+_17912834 | 0.18 |
ENSDART00000104647
|
prim1
|
primase, DNA, polypeptide 1 |
| chr23_-_5849341 | 0.17 |
ENSDART00000019455
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr11_+_6450876 | 0.17 |
ENSDART00000170180
|
CU062501.2
|
ENSDARG00000103967 |
| chr17_-_5453436 | 0.17 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr6_+_58528350 | 0.16 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr3_-_36143057 | 0.15 |
ENSDART00000141638
|
prkar1aa
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a |
| chr11_+_42438405 | 0.14 |
ENSDART00000163269
|
zgc:194981
|
zgc:194981 |
| chr25_+_35540123 | 0.13 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr14_-_33180680 | 0.13 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr1_-_22679567 | 0.12 |
ENSDART00000126785
|
pds5a
|
PDS5 cohesin associated factor A |
| chr1_+_45148233 | 0.12 |
ENSDART00000048191
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr22_+_30386668 | 0.12 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr6_-_55411751 | 0.11 |
ENSDART00000159246
ENSDART00000158929 |
ctsa
|
cathepsin A |
| chr23_-_21520379 | 0.10 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr25_+_34410545 | 0.10 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr20_+_31372117 | 0.09 |
|
|
|
| chr3_-_4648706 | 0.09 |
|
|
|
| chr13_-_35716712 | 0.08 |
ENSDART00000170106
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr11_-_26463925 | 0.08 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr18_-_46371631 | 0.07 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr3_+_32731002 | 0.07 |
ENSDART00000137599
|
zgc:162613
|
zgc:162613 |
| chr8_+_47815153 | 0.06 |
ENSDART00000053285
|
ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 |
| chr24_-_37989711 | 0.03 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr12_+_25132396 | 0.03 |
ENSDART00000139362
|
mta3
|
metastasis associated 1 family, member 3 |
| chr10_+_572931 | 0.02 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr19_+_40661954 | 0.01 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.4 | 1.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 3.6 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 4.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 3.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) regulation of neutrophil migration(GO:1902622) |
| 0.2 | 3.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.5 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 3.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 2.4 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.3 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.7 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.0 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.5 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 3.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) terpenoid biosynthetic process(GO:0016114) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.9 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.3 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 2.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 2.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.6 | GO:1990077 | primosome complex(GO:1990077) |
| 0.2 | 3.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 3.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.9 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 1.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 4.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.2 | 0.6 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.2 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 3.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.4 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 3.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.0 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 4.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 1.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 3.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |