Project
DANIO-CODE
Navigation
Downloads
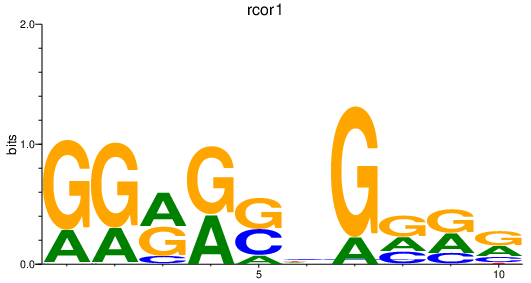
Results for rcor1
Z-value: 0.95
Transcription factors associated with rcor1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rcor1
|
ENSDARG00000031434 | REST corepressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rcor1 | dr10_dc_chr17_-_29253918_29253959 | -0.81 | 1.7e-04 | Click! |
Activity profile of rcor1 motif
Sorted Z-values of rcor1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rcor1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_182037 | 3.48 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr15_-_33816 | 3.43 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr12_-_4351924 | 2.85 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr3_-_6078015 | 2.45 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr3_-_32686790 | 2.24 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr3_+_59560881 | 1.90 |
ENSDART00000084738
|
ppp1r27a
|
protein phosphatase 1, regulatory subunit 27a |
| chr3_-_60929921 | 1.87 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr1_-_9387290 | 1.77 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr7_+_50828119 | 1.75 |
ENSDART00000108738
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr12_-_4351401 | 1.74 |
|
|
|
| chr5_-_1698166 | 1.66 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr7_-_33558939 | 1.66 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr2_+_49837788 | 1.62 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr1_-_25272318 | 1.60 |
ENSDART00000134192
|
synpo2b
|
synaptopodin 2b |
| chr23_+_45020761 | 1.50 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr16_+_7213161 | 1.50 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr12_-_4033069 | 1.50 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr19_-_2399429 | 1.45 |
ENSDART00000043595
|
twist1a
|
twist family bHLH transcription factor 1a |
| chr11_+_14618598 | 1.44 |
ENSDART00000109308
|
si:ch73-60h1.1
|
si:ch73-60h1.1 |
| chr22_+_24131239 | 1.43 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr3_+_30790513 | 1.41 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr23_-_7283514 | 1.40 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr4_+_21485280 | 1.38 |
ENSDART00000066896
|
syt1a
|
synaptotagmin Ia |
| chr4_-_12863235 | 1.38 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| KN150640v1_+_5728 | 1.37 |
|
|
|
| chr13_-_28177612 | 1.31 |
ENSDART00000045351
|
lbx1a
|
ladybird homeobox 1a |
| chr10_-_27261937 | 1.28 |
|
|
|
| chr10_+_9052152 | 1.28 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr14_-_9114872 | 1.28 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr8_-_38357549 | 1.25 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| KN149707v1_+_4014 | 1.25 |
ENSDART00000168516
|
cdo1
|
cysteine dioxygenase, type I |
| chr22_-_951880 | 1.25 |
ENSDART00000105895
ENSDART00000172206 |
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr19_+_42183946 | 1.23 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr9_-_53220424 | 1.22 |
|
|
|
| chr2_-_55971357 | 1.21 |
ENSDART00000154701
ENSDART00000154107 |
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr4_-_876235 | 1.19 |
|
|
|
| chr23_+_14461509 | 1.19 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr8_+_13465718 | 1.17 |
ENSDART00000034740
|
fut9d
|
fucosyltransferase 9d |
| chr22_+_10172134 | 1.16 |
ENSDART00000132641
|
pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr16_-_24602919 | 1.15 |
ENSDART00000147478
|
cadm4
|
cell adhesion molecule 4 |
| chr22_+_30060679 | 1.14 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr3_-_60930025 | 1.13 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr4_-_12862869 | 1.12 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr8_+_15968469 | 1.11 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_-_59013059 | 1.11 |
|
|
|
| chr21_-_36338704 | 1.09 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr11_-_32461160 | 1.07 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr7_+_16583234 | 1.07 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr4_+_14972481 | 1.06 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr23_+_29962550 | 1.06 |
ENSDART00000149378
|
mxra8b
|
matrix-remodelling associated 8b |
| chr14_-_549816 | 1.05 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr7_-_33558771 | 1.05 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr21_-_36338514 | 1.04 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr18_+_5456221 | 1.04 |
|
|
|
| chr9_-_22371512 | 1.04 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr1_-_14642681 | 1.04 |
ENSDART00000109970
|
dlc1
|
DLC1 Rho GTPase activating protein |
| chr10_+_10830549 | 1.04 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr1_+_18842158 | 1.03 |
ENSDART00000103089
|
ENSDARG00000070320
|
ENSDARG00000070320 |
| chr12_+_16403743 | 1.02 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr1_-_18110990 | 1.02 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr4_+_11312432 | 1.00 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr7_+_58384379 | 1.00 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr24_-_14567403 | 0.98 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr20_-_158899 | 0.98 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr16_-_10089440 | 0.98 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr6_+_58549236 | 0.96 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr5_-_43896815 | 0.96 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr1_+_44660098 | 0.96 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr22_+_24131297 | 0.96 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_+_11902170 | 0.95 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr8_-_38308535 | 0.95 |
|
|
|
| chr16_+_12101666 | 0.94 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr11_-_13069266 | 0.92 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr19_+_17941678 | 0.91 |
ENSDART00000124597
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr7_-_52848084 | 0.90 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr1_-_21987718 | 0.90 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr1_-_30298698 | 0.89 |
|
|
|
| chr20_+_23339698 | 0.89 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr6_+_251026 | 0.88 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr12_-_10372055 | 0.87 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr10_+_9575 | 0.87 |
|
|
|
| chr4_-_19027117 | 0.86 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr4_+_9393501 | 0.83 |
ENSDART00000092013
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr4_+_14972708 | 0.83 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr1_+_44044807 | 0.83 |
ENSDART00000133926
ENSDART00000049701 |
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr12_+_30471689 | 0.82 |
ENSDART00000124920
|
nrap
|
nebulin-related anchoring protein |
| chr14_-_43191278 | 0.81 |
ENSDART00000169615
|
adh5
|
alcohol dehydrogenase 5 |
| chr9_-_10561062 | 0.80 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr24_+_30343717 | 0.79 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr16_-_34304851 | 0.79 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr21_+_44586326 | 0.79 |
|
|
|
| chr16_+_29109050 | 0.78 |
ENSDART00000122681
|
CR848841.1
|
Nestin |
| chr18_+_40364675 | 0.78 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr6_-_31246730 | 0.78 |
ENSDART00000079173
|
lepr
|
leptin receptor |
| chr5_-_71505131 | 0.77 |
ENSDART00000037691
|
tbx5a
|
T-box 5a |
| chr3_+_27684309 | 0.77 |
|
|
|
| chr1_+_54403123 | 0.76 |
ENSDART00000152654
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr5_-_43896757 | 0.76 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr20_-_25727145 | 0.76 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr3_+_16080070 | 0.76 |
ENSDART00000137396
|
rpl19
|
ribosomal protein L19 |
| chr19_-_15951003 | 0.76 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr19_+_14197118 | 0.75 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr16_+_10974655 | 0.74 |
|
|
|
| chr8_+_20125687 | 0.74 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr19_-_28382815 | 0.74 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr19_+_32679258 | 0.74 |
|
|
|
| chr8_-_15071283 | 0.74 |
|
|
|
| chr11_-_44280978 | 0.73 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr13_-_40190349 | 0.73 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr2_+_37985514 | 0.72 |
ENSDART00000149224
|
apol1
|
apolipoprotein L, 1 |
| chr13_-_219661 | 0.72 |
ENSDART00000133731
|
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr4_+_13932849 | 0.72 |
ENSDART00000141742
|
pphln1
|
periphilin 1 |
| chr11_-_1792616 | 0.71 |
|
|
|
| chr14_+_34207218 | 0.71 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| KN150361v1_-_15974 | 0.70 |
ENSDART00000167588
|
ENSDARG00000099568
|
ENSDARG00000099568 |
| chr19_-_34508557 | 0.70 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr20_+_1724609 | 0.69 |
|
|
|
| chr19_+_32679320 | 0.69 |
|
|
|
| chr12_+_5495284 | 0.69 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr16_-_21981065 | 0.69 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr25_-_255242 | 0.68 |
|
|
|
| chr20_+_27748136 | 0.68 |
ENSDART00000141697
|
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr21_+_19889712 | 0.66 |
ENSDART00000147294
|
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr6_+_9185750 | 0.66 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr6_+_41257803 | 0.65 |
ENSDART00000042683
|
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr18_+_36825511 | 0.65 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr7_-_33559032 | 0.63 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr6_-_16156494 | 0.63 |
|
|
|
| chr14_-_2278018 | 0.62 |
ENSDART00000125674
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr8_-_26848577 | 0.62 |
|
|
|
| chr7_+_50828247 | 0.60 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr9_+_21725007 | 0.60 |
ENSDART00000045093
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr17_+_48999061 | 0.59 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr19_-_28192439 | 0.59 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr7_-_26331734 | 0.59 |
|
|
|
| chr22_-_16971035 | 0.58 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr20_-_54645287 | 0.57 |
ENSDART00000153389
|
yy1b
|
YY1 transcription factor b |
| chr24_+_26944182 | 0.57 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr25_-_34638406 | 0.56 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr18_-_11426 | 0.56 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr2_-_39052185 | 0.56 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr16_-_25370472 | 0.55 |
ENSDART00000154543
|
prelid3a
|
PRELI domain containing 3A |
| chr13_+_28382422 | 0.55 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr6_-_60109646 | 0.55 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr10_+_17490443 | 0.54 |
|
|
|
| chr6_+_37881015 | 0.52 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr19_+_42491631 | 0.52 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr17_-_37040941 | 0.51 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr17_-_120568 | 0.51 |
|
|
|
| chr18_+_19783732 | 0.51 |
ENSDART00000043455
|
smad3b
|
SMAD family member 3b |
| chr22_-_16970979 | 0.50 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr12_-_33792377 | 0.50 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr3_+_33209227 | 0.50 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr15_-_33782909 | 0.49 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr1_+_44239727 | 0.49 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr17_-_120755 | 0.49 |
|
|
|
| chr24_+_30343687 | 0.49 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr12_-_3320438 | 0.49 |
|
|
|
| chr23_+_33457343 | 0.48 |
ENSDART00000114423
|
rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_-_117828 | 0.47 |
|
|
|
| chr15_-_16271208 | 0.47 |
|
|
|
| chr17_+_11961595 | 0.47 |
|
|
|
| chr14_-_35985200 | 0.46 |
ENSDART00000172783
|
egf
|
epidermal growth factor |
| chr15_+_29190162 | 0.46 |
ENSDART00000105222
|
zgc:101731
|
zgc:101731 |
| chr22_-_16971180 | 0.46 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr14_-_9274939 | 0.46 |
|
|
|
| chr12_+_272930 | 0.46 |
ENSDART00000152639
|
ict1
|
immature colon carcinoma transcript 1 |
| chr12_-_21918058 | 0.46 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr8_+_22559432 | 0.46 |
|
|
|
| chr4_-_16344835 | 0.45 |
ENSDART00000134449
|
epyc
|
epiphycan |
| chr5_+_22173976 | 0.45 |
ENSDART00000142112
|
vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr3_+_22447812 | 0.44 |
ENSDART00000112270
|
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr3_+_28371725 | 0.43 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr4_-_12863112 | 0.43 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr8_+_7655655 | 0.43 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr8_+_30654835 | 0.43 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr1_+_52308366 | 0.43 |
ENSDART00000108492
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr11_-_16067646 | 0.43 |
|
|
|
| chr9_-_55878647 | 0.42 |
|
|
|
| chr14_-_2451650 | 0.41 |
ENSDART00000111748
|
pcdhb
|
protocadherin b |
| chr12_+_3686860 | 0.41 |
ENSDART00000152482
|
pagr1
|
PAXIP1 associated glutamate-rich protein 1 |
| KN150640v1_+_5907 | 0.41 |
|
|
|
| chr13_-_24248675 | 0.40 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr21_+_5293086 | 0.40 |
|
|
|
| chr5_-_44243476 | 0.40 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr11_+_44922098 | 0.40 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr23_+_22408855 | 0.39 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr15_+_9351211 | 0.39 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr12_-_31342432 | 0.39 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr16_-_20084451 | 0.39 |
ENSDART00000079155
|
hdac9b
|
histone deacetylase 9b |
| chr2_-_44429891 | 0.39 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr10_-_29386085 | 0.39 |
ENSDART00000022605
|
tmem126a
|
transmembrane protein 126A |
| chr17_+_42003279 | 0.38 |
ENSDART00000111537
|
kiz
|
kizuna centrosomal protein |
| chr18_+_45199548 | 0.38 |
|
|
|
| chr2_+_38244259 | 0.38 |
ENSDART00000131384
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr24_-_25021505 | 0.37 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| KN149959v1_+_21015 | 0.37 |
ENSDART00000161495
|
CABZ01078608.1
|
ENSDARG00000105028 |
| chr14_-_30467807 | 0.37 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr11_+_14373731 | 0.37 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr7_-_24119645 | 0.37 |
ENSDART00000141769
|
ptgr1
|
prostaglandin reductase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0034433 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) reverse cholesterol transport(GO:0043691) positive regulation of steroid metabolic process(GO:0045940) |
| 0.6 | 3.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 1.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.4 | 1.9 | GO:0048745 | smooth muscle tissue development(GO:0048745) arterial endothelial cell fate commitment(GO:0060844) |
| 0.3 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.6 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 2.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 0.8 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.2 | 0.7 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 0.7 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.2 | 1.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 0.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 1.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 1.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 0.7 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.2 | 0.9 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.2 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 2.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 0.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.9 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.0 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.1 | 0.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.4 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.6 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.0 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.1 | 0.4 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 1.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.6 | GO:0061469 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 1.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.7 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.0 | 1.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.9 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 1.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.2 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.9 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 1.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0060282 | cGMP-mediated signaling(GO:0019934) positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 1.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.8 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.5 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.6 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 1.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 0.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.5 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 6.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.3 | 0.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.9 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.0 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.2 | 2.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.8 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.8 | GO:0035381 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 3.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.6 | GO:0035586 | G-protein coupled adenosine receptor activity(GO:0001609) purinergic receptor activity(GO:0035586) |
| 0.1 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.2 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 3.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 8.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 1.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |