Project
DANIO-CODE
Navigation
Downloads
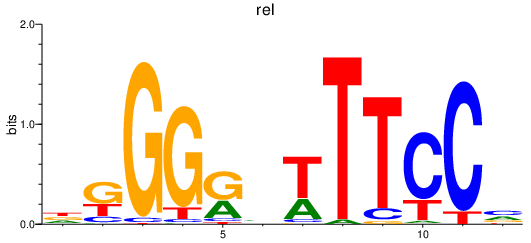
Results for rel
Z-value: 0.28
Transcription factors associated with rel
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rel
|
ENSDARG00000055276 | v-rel avian reticuloendotheliosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rel | dr10_dc_chr13_-_25689369_25689385 | -0.39 | 1.4e-01 | Click! |
Activity profile of rel motif
Sorted Z-values of rel motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rel
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_33817169 | 0.47 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr11_+_7148830 | 0.41 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr2_+_42342148 | 0.40 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr22_-_613948 | 0.38 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr20_-_49880744 | 0.35 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr20_-_44598129 | 0.31 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr2_+_17112049 | 0.28 |
ENSDART00000125647
|
tfa
|
transferrin-a |
| chr24_-_33817036 | 0.27 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr8_+_47644421 | 0.26 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr16_+_33041823 | 0.25 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr5_+_36431824 | 0.22 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_53968095 | 0.21 |
ENSDART00000162689
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr16_+_43249142 | 0.21 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr13_-_39821399 | 0.21 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr13_+_3533517 | 0.19 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr7_-_58649991 | 0.19 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr24_+_9073790 | 0.19 |
|
|
|
| chr15_+_17164373 | 0.19 |
ENSDART00000123197
ENSDART00000154418 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr15_+_17164535 | 0.18 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_-_22590638 | 0.18 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_+_3533318 | 0.18 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr6_+_59788008 | 0.18 |
|
|
|
| chr13_+_3533426 | 0.17 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr3_-_32202533 | 0.17 |
ENSDART00000155757
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr25_+_5695630 | 0.17 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr3_+_23587156 | 0.16 |
|
|
|
| chr7_+_57423271 | 0.16 |
ENSDART00000056466
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr3_-_42949225 | 0.16 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr7_-_39170016 | 0.16 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr6_+_44817077 | 0.16 |
ENSDART00000169713
|
chl1b
|
cell adhesion molecule L1-like b |
| chr10_+_43950311 | 0.15 |
ENSDART00000027242
|
nr2f1b
|
nuclear receptor subfamily 2, group F, member 1b |
| chr14_-_25279835 | 0.15 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr23_-_34970838 | 0.15 |
ENSDART00000087219
|
ENSDARG00000061272
|
ENSDARG00000061272 |
| chr13_-_4238224 | 0.15 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr16_-_6318609 | 0.15 |
|
|
|
| chr8_+_7739964 | 0.14 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr3_+_54489642 | 0.14 |
ENSDART00000114443
|
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr24_-_14567403 | 0.13 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr4_-_16344954 | 0.13 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr21_-_43915591 | 0.13 |
|
|
|
| chr24_-_12543205 | 0.13 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr24_+_4341242 | 0.13 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr18_+_30868972 | 0.13 |
ENSDART00000176162
|
foxf1
|
forkhead box F1 |
| chr21_+_22808694 | 0.13 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr13_+_14873339 | 0.13 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr6_+_33556155 | 0.13 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr6_+_33556031 | 0.12 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr23_-_3760604 | 0.12 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr12_-_19129006 | 0.12 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr8_+_7740132 | 0.12 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_+_36545240 | 0.12 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr3_+_10460077 | 0.11 |
ENSDART00000122844
|
TMEM100
|
transmembrane protein 100 |
| chr24_-_38928988 | 0.11 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr6_-_35326559 | 0.11 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr10_-_11806373 | 0.11 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr4_+_119964 | 0.11 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr20_+_230164 | 0.10 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr12_-_34786844 | 0.10 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr22_+_31866783 | 0.10 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr15_+_6462607 | 0.10 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr8_-_46313975 | 0.10 |
ENSDART00000075189
|
mtor
|
mechanistic target of rapamycin (serine/threonine kinase) |
| chr3_-_21111908 | 0.10 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr15_-_27431216 | 0.09 |
ENSDART00000155933
|
AL954322.2
|
ENSDARG00000097072 |
| chr9_+_21581061 | 0.09 |
|
|
|
| chr16_+_13964949 | 0.09 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| KN150630v1_+_44324 | 0.09 |
|
|
|
| chr14_+_20595818 | 0.09 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr25_+_5162045 | 0.09 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr23_-_36319185 | 0.08 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr14_+_26498814 | 0.08 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr16_+_43249077 | 0.08 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr16_+_1227270 | 0.08 |
|
|
|
| chr19_-_7151715 | 0.08 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr23_+_43531865 | 0.08 |
ENSDART00000022498
|
tti1
|
TELO2 interacting protein 1 |
| chr8_+_47908640 | 0.08 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr21_-_36495488 | 0.08 |
|
|
|
| chr19_+_7798512 | 0.08 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr25_+_5695588 | 0.08 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr13_-_36400356 | 0.08 |
ENSDART00000175311
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr22_-_16153692 | 0.08 |
ENSDART00000171331
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr2_-_10555152 | 0.08 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr13_+_10100146 | 0.08 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr24_-_36144354 | 0.08 |
|
|
|
| chr20_-_49880706 | 0.08 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr23_+_758658 | 0.07 |
|
|
|
| chr17_+_8018808 | 0.07 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr3_+_34540552 | 0.07 |
ENSDART00000007073
ENSDART00000133457 |
dlx4a
|
distal-less homeobox 4a |
| chr23_+_26995808 | 0.07 |
|
|
|
| chr9_+_1225808 | 0.07 |
ENSDART00000177730
|
CABZ01114359.1
|
ENSDARG00000107255 |
| chr20_-_34074096 | 0.07 |
ENSDART00000136834
|
sele
|
selectin E |
| chr6_-_31246730 | 0.07 |
ENSDART00000079173
|
lepr
|
leptin receptor |
| chr16_+_29696109 | 0.07 |
ENSDART00000153683
|
VPS72
|
vacuolar protein sorting 72 homolog |
| chr21_-_30508374 | 0.07 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr8_+_39640881 | 0.07 |
ENSDART00000040330
ENSDART00000045529 ENSDART00000159852 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr15_-_33782909 | 0.07 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr10_+_18919823 | 0.07 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr11_+_2211050 | 0.07 |
ENSDART00000123944
|
hoxc13b
|
homeobox C13b |
| chr9_-_28464236 | 0.06 |
ENSDART00000146932
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr2_-_49060868 | 0.06 |
|
|
|
| chr13_+_1051015 | 0.06 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_67508752 | 0.06 |
ENSDART00000167891
|
CR388184.1
|
ENSDARG00000099896 |
| chr19_+_48692152 | 0.06 |
ENSDART00000169729
|
sgol1
|
shugoshin-like 1 (S. pombe) |
| chr2_-_28687065 | 0.06 |
ENSDART00000164657
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr19_-_35828807 | 0.06 |
|
|
|
| chr8_-_17580655 | 0.06 |
|
|
|
| chr15_+_37534857 | 0.06 |
ENSDART00000124779
|
igflr1
|
IGF-like family receptor 1 |
| chr18_-_18554151 | 0.06 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr11_-_33925792 | 0.06 |
ENSDART00000172978
|
atp13a3
|
ATPase type 13A3 |
| chr13_-_22713071 | 0.06 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr15_+_32963516 | 0.06 |
ENSDART00000167739
|
dclk1b
|
doublecortin-like kinase 1b |
| chr2_+_2583068 | 0.06 |
|
|
|
| chr18_-_16337647 | 0.06 |
ENSDART00000110278
|
rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr3_+_19066717 | 0.05 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr12_-_34786663 | 0.05 |
ENSDART00000158848
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr9_+_21524891 | 0.05 |
|
|
|
| chr17_-_29232289 | 0.05 |
ENSDART00000031458
|
traf3
|
TNF receptor-associated factor 3 |
| chr3_+_35366133 | 0.05 |
ENSDART00000164420
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr24_+_39201883 | 0.05 |
|
|
|
| chr15_+_32963784 | 0.05 |
ENSDART00000167739
|
dclk1b
|
doublecortin-like kinase 1b |
| chr17_-_23235070 | 0.05 |
|
|
|
| chr23_+_44951868 | 0.05 |
|
|
|
| chr16_-_2972770 | 0.05 |
ENSDART00000082522
|
abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr15_-_9655500 | 0.04 |
ENSDART00000171214
ENSDART00000171911 |
nars2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr24_-_35486496 | 0.04 |
|
|
|
| chr1_+_44516242 | 0.04 |
ENSDART00000074683
|
EVI5L
|
ecotropic viral integration site 5 like |
| chr8_+_25984588 | 0.04 |
|
|
|
| chr25_-_28483417 | 0.04 |
ENSDART00000112850
|
mettl20
|
methyltransferase like 20 |
| chr12_-_26339002 | 0.04 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr6_-_19556328 | 0.04 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr2_-_3202918 | 0.04 |
|
|
|
| chr6_+_37677155 | 0.04 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr7_-_50546498 | 0.03 |
|
|
|
| chr15_+_6462447 | 0.03 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr7_+_1228387 | 0.03 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr9_+_3147601 | 0.03 |
ENSDART00000135619
|
ftr52p
|
finTRIM family, member 52, pseudogene |
| chr10_-_321050 | 0.03 |
ENSDART00000165244
ENSDART00000161493 |
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr12_-_34787076 | 0.03 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr7_+_56171081 | 0.03 |
ENSDART00000073596
ENSDART00000123273 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr6_+_23462412 | 0.03 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr25_+_4729095 | 0.03 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr8_-_18637019 | 0.03 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr15_-_16241500 | 0.03 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr11_-_19414624 | 0.03 |
ENSDART00000157847
|
atxn7
|
ataxin 7 |
| chr19_-_7151801 | 0.02 |
ENSDART00000176808
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr9_+_24344001 | 0.02 |
|
|
|
| chr18_-_11426 | 0.02 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr23_+_3570270 | 0.02 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr5_-_31306608 | 0.02 |
|
|
|
| chr16_+_40625705 | 0.02 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr14_+_6639864 | 0.02 |
ENSDART00000061001
|
gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr6_-_52677210 | 0.02 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr15_+_27431469 | 0.02 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr9_+_492726 | 0.02 |
ENSDART00000112635
|
ENSDARG00000078172
|
ENSDARG00000078172 |
| chr14_-_30556788 | 0.02 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr2_-_7772932 | 0.02 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr5_+_29988157 | 0.01 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr11_-_13295436 | 0.01 |
|
|
|
| chr17_+_48999061 | 0.01 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr7_+_34923319 | 0.01 |
|
|
|
| chr14_+_46652294 | 0.01 |
|
|
|
| chr2_-_29501573 | 0.01 |
ENSDART00000138073
|
cahz
|
carbonic anhydrase |
| chr15_+_20961370 | 0.01 |
ENSDART00000154854
|
CR848784.5
|
ENSDARG00000097996 |
| chr11_+_40280114 | 0.01 |
ENSDART00000175424
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr18_-_25918992 | 0.01 |
ENSDART00000144145
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr22_-_31722970 | 0.01 |
ENSDART00000128247
|
lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_28167936 | 0.01 |
ENSDART00000043466
|
traf2a
|
Tnf receptor-associated factor 2a |
| chr8_+_22334642 | 0.01 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr16_+_24766043 | 0.01 |
ENSDART00000114304
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr21_-_44013995 | 0.01 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr13_-_25066650 | 0.01 |
|
|
|
| chr24_+_33478309 | 0.01 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr8_+_48614626 | 0.01 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr14_+_14501263 | 0.01 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr13_+_51154848 | 0.01 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr22_+_8506974 | 0.00 |
ENSDART00000132998
|
si:ch73-27e22.7
|
si:ch73-27e22.7 |
| chr3_+_23587109 | 0.00 |
|
|
|
| chr9_-_34145854 | 0.00 |
|
|
|
| chr3_-_31487744 | 0.00 |
ENSDART00000124559
|
moto
|
minamoto |
| chr23_-_43531791 | 0.00 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr21_-_2322963 | 0.00 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr12_+_27370834 | 0.00 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr15_-_44054313 | 0.00 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr3_-_42949449 | 0.00 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr20_-_29961621 | 0.00 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr15_-_8343858 | 0.00 |
ENSDART00000137552
|
si:ch211-113p18.3
|
si:ch211-113p18.3 |
| chr16_+_10666906 | 0.00 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr6_+_3703160 | 0.00 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr1_+_11448327 | 0.00 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.3 | GO:0019731 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.1 | 0.8 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.1 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |