Project
DANIO-CODE
Navigation
Downloads
Results for rela_nfkb1
Z-value: 0.66
Transcription factors associated with rela_nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rela
|
ENSDARG00000098696 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
|
nfkb1
|
ENSDARG00000105261 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rela | dr10_dc_chr7_-_667744_667811 | 0.51 | 4.4e-02 | Click! |
| nfkb1 | dr10_dc_chr14_-_50657786_50657854 | 0.18 | 5.1e-01 | Click! |
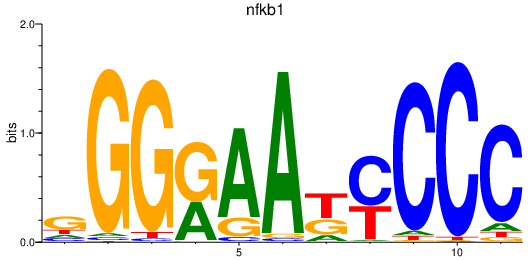
Activity profile of rela_nfkb1 motif
Sorted Z-values of rela_nfkb1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rela_nfkb1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_16934961 | 1.30 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr11_+_6446302 | 1.29 |
ENSDART00000140707
ENSDART00000036939 |
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr11_+_34788205 | 1.24 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr20_-_35173530 | 1.00 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr8_-_1669966 | 0.95 |
|
|
|
| chr23_+_10412852 | 0.88 |
ENSDART00000142595
|
krt18
|
keratin 18 |
| chr12_-_30645906 | 0.84 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr11_-_19414624 | 0.81 |
ENSDART00000157847
|
atxn7
|
ataxin 7 |
| chr16_+_4288700 | 0.81 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| KN149832v1_+_6032 | 0.80 |
|
|
|
| chr2_+_17112049 | 0.78 |
ENSDART00000125647
|
tfa
|
transferrin-a |
| chr5_+_50355421 | 0.74 |
ENSDART00000074638
|
ankrd31
|
ankyrin repeat domain 31 |
| chr9_-_26776407 | 0.70 |
|
|
|
| chr7_+_1228387 | 0.69 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr20_+_16982314 | 0.68 |
ENSDART00000146625
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr5_+_36431907 | 0.68 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr22_+_30234718 | 0.68 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| KN150670v1_-_72156 | 0.67 |
|
|
|
| chr3_-_26393945 | 0.66 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr12_+_34631310 | 0.65 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr2_-_51231808 | 0.64 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr12_+_34631232 | 0.61 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr21_-_485916 | 0.59 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr25_+_24520476 | 0.58 |
|
|
|
| chr15_+_27431469 | 0.58 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr1_+_9426040 | 0.57 |
ENSDART00000080576
|
lratb
|
lecithin retinol acyltransferase b (phosphatidylcholine-retinol O-acyltransferase) |
| chr13_-_33696425 | 0.56 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr4_-_25496215 | 0.56 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_-_667744 | 0.52 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr3_-_24925189 | 0.52 |
ENSDART00000156459
|
ep300b
|
E1A binding protein p300 b |
| chr13_+_1051015 | 0.51 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_-_18839556 | 0.51 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr7_-_48392300 | 0.51 |
|
|
|
| chr3_+_22412059 | 0.50 |
|
|
|
| chr8_+_22334642 | 0.50 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr7_+_1228148 | 0.49 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr4_+_29209923 | 0.49 |
ENSDART00000168709
|
ENSDARG00000104041
|
ENSDARG00000104041 |
| chr16_+_1227270 | 0.47 |
|
|
|
| chr20_-_21775455 | 0.45 |
|
|
|
| chr11_-_5868257 | 0.44 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr3_+_35276575 | 0.43 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr4_-_12796590 | 0.42 |
ENSDART00000075127
|
b2m
|
beta-2-microglobulin |
| chr11_+_8558222 | 0.41 |
ENSDART00000169141
ENSDART00000126523 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr8_+_28240085 | 0.41 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr7_+_15623852 | 0.41 |
ENSDART00000161608
|
pax6b
|
paired box 6b |
| chr14_-_30556788 | 0.39 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr11_-_7129695 | 0.39 |
ENSDART00000016472
|
bmp7a
|
bone morphogenetic protein 7a |
| chr3_-_10342940 | 0.39 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr8_+_7740132 | 0.38 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr16_+_24766043 | 0.37 |
ENSDART00000114304
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr16_+_49796978 | 0.37 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr3_+_54876680 | 0.35 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr6_+_1895734 | 0.35 |
ENSDART00000109679
|
quo
|
quattro |
| chr13_-_35908743 | 0.34 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr7_-_46509291 | 0.34 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr2_-_39709905 | 0.33 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr13_-_24181106 | 0.33 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr2_+_58658615 | 0.33 |
|
|
|
| chr19_-_11290766 | 0.33 |
ENSDART00000044426
|
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr9_+_55999688 | 0.33 |
ENSDART00000172063
|
edar
|
ectodysplasin A receptor |
| chr23_+_6043862 | 0.33 |
|
|
|
| chr17_+_48999061 | 0.32 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr11_+_26138359 | 0.32 |
ENSDART00000087652
|
cpne1
|
copine I |
| chr2_-_3202918 | 0.31 |
|
|
|
| chr4_+_119964 | 0.30 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr6_+_58921655 | 0.30 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr23_-_2084541 | 0.30 |
ENSDART00000091532
|
ndnf
|
neuron-derived neurotrophic factor |
| chr1_+_44137973 | 0.29 |
|
|
|
| chr5_-_43896815 | 0.29 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr21_+_28921734 | 0.29 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_25106923 | 0.29 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr22_-_20316908 | 0.29 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr11_+_30049795 | 0.28 |
|
|
|
| chr24_-_15313198 | 0.28 |
|
|
|
| chr16_-_31835463 | 0.28 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr19_+_43524566 | 0.27 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr5_+_23582396 | 0.27 |
ENSDART00000051549
ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr17_+_1950129 | 0.27 |
ENSDART00000110529
|
bub1bb
|
BUB1 mitotic checkpoint serine/threonine kinase Bb |
| chr14_-_4014025 | 0.27 |
ENSDART00000157782
|
irf2
|
interferon regulatory factor 2 |
| chr25_+_5162045 | 0.27 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr22_+_470536 | 0.26 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr12_+_36796886 | 0.26 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr21_+_30900987 | 0.25 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr15_-_6979997 | 0.25 |
ENSDART00000169944
|
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr24_+_34227202 | 0.25 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| KN150680v1_+_10854 | 0.25 |
|
|
|
| chr16_+_10666588 | 0.25 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr13_-_4238408 | 0.24 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr18_-_11707 | 0.24 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr8_-_38444845 | 0.24 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr16_-_16682692 | 0.24 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr17_+_48999102 | 0.24 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr7_+_15623685 | 0.24 |
ENSDART00000146704
|
pax6b
|
paired box 6b |
| chr20_-_35173500 | 0.23 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr24_-_4941954 | 0.23 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr14_-_30739765 | 0.23 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr6_+_33556031 | 0.23 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr2_+_44067425 | 0.22 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr20_-_6153133 | 0.22 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr6_+_3703684 | 0.22 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr22_-_613948 | 0.22 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr12_+_27370834 | 0.21 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr16_-_36022327 | 0.21 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr19_-_35828807 | 0.21 |
|
|
|
| chr18_-_18554151 | 0.21 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr25_+_3221696 | 0.21 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr2_+_2583068 | 0.21 |
|
|
|
| chr9_-_34129128 | 0.21 |
|
|
|
| chr15_-_33782909 | 0.20 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr2_-_3203802 | 0.20 |
ENSDART00000105818
|
guk1a
|
guanylate kinase 1a |
| chr7_+_40183670 | 0.20 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr7_+_40879017 | 0.20 |
|
|
|
| chr13_+_14873339 | 0.20 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr8_+_42992323 | 0.20 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr7_-_38590391 | 0.20 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr8_+_25984588 | 0.20 |
|
|
|
| chr14_+_38506051 | 0.19 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr5_+_20596549 | 0.19 |
ENSDART00000051586
|
hs3st1l2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1-like 2 |
| chr14_+_38505790 | 0.19 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr11_-_35501227 | 0.19 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr3_-_7769397 | 0.18 |
|
|
|
| chr15_+_24821561 | 0.18 |
ENSDART00000156424
|
cpda
|
carboxypeptidase D, a |
| chr21_-_30378764 | 0.18 |
ENSDART00000136247
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr5_+_36431824 | 0.18 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr6_-_53333648 | 0.18 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr16_+_13964949 | 0.18 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr11_+_40280114 | 0.18 |
ENSDART00000175424
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr4_+_13811932 | 0.18 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr13_-_35908796 | 0.18 |
ENSDART00000084929
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr5_+_10789834 | 0.18 |
|
|
|
| chr2_-_42011586 | 0.18 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr4_+_2210362 | 0.18 |
ENSDART00000067427
|
yeats4
|
YEATS domain containing 4 |
| chr5_-_43896757 | 0.17 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr6_-_35326559 | 0.17 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr7_+_16583234 | 0.17 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr14_+_150214 | 0.17 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr3_+_23587156 | 0.17 |
|
|
|
| chr13_+_35213326 | 0.17 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| KN150183v1_+_12947 | 0.17 |
|
|
|
| chr23_+_19775308 | 0.17 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr18_+_22285979 | 0.17 |
|
|
|
| chr2_+_42342148 | 0.17 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr7_+_61770242 | 0.17 |
ENSDART00000092580
|
rbpjb
|
recombination signal binding protein for immunoglobulin kappa J region b |
| chr7_+_39487676 | 0.17 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr10_+_10393377 | 0.16 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr8_+_53390978 | 0.16 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr16_-_13914368 | 0.16 |
|
|
|
| chr20_-_51494625 | 0.16 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr7_-_46509218 | 0.15 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr21_+_3556841 | 0.15 |
|
|
|
| chr21_-_44013995 | 0.15 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr18_+_730277 | 0.15 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr16_+_12740993 | 0.15 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr25_+_16549708 | 0.14 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr9_+_13762263 | 0.14 |
ENSDART00000141314
ENSDART00000138254 |
abi2a
|
abl-interactor 2a |
| chr18_-_15132837 | 0.14 |
ENSDART00000172273
|
btbd11b
|
BTB (POZ) domain containing 11b |
| chr7_-_73620490 | 0.14 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr3_+_22411884 | 0.14 |
|
|
|
| chr19_-_32913075 | 0.14 |
ENSDART00000052104
|
fuca1.1
|
fucosidase, alpha-L- 1, tissue, tandem duplicate 1 |
| chr11_-_32502472 | 0.14 |
|
|
|
| chr15_-_27431216 | 0.14 |
ENSDART00000155933
|
AL954322.2
|
ENSDARG00000097072 |
| KN149808v1_+_14146 | 0.14 |
|
|
|
| chr4_-_2209924 | 0.14 |
ENSDART00000082289
|
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr2_+_25107108 | 0.14 |
ENSDART00000136072
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr12_-_13690508 | 0.14 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr17_+_9918253 | 0.14 |
ENSDART00000169892
|
srp54
|
signal recognition particle 54 |
| chr6_-_53333532 | 0.14 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr3_+_35277555 | 0.13 |
|
|
|
| chr3_+_48362748 | 0.13 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr16_+_33041823 | 0.13 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr1_+_11448327 | 0.13 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr16_+_26892979 | 0.13 |
ENSDART00000048036
|
gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr23_-_36319185 | 0.13 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr19_+_20194594 | 0.13 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| KN150437v1_-_892 | 0.13 |
ENSDART00000178017
ENSDART00000175804 |
CABZ01103416.1
|
ENSDARG00000106284 |
| chr21_+_22808694 | 0.12 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr13_+_3533517 | 0.12 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr17_+_38314814 | 0.12 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr3_+_59608335 | 0.12 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr12_-_36145252 | 0.12 |
|
|
|
| chr7_+_19583578 | 0.12 |
ENSDART00000149812
|
ovol1
|
ovo-like zinc finger 1 |
| chr16_+_10666906 | 0.12 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr8_+_5222065 | 0.12 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr11_+_18710724 | 0.12 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr11_-_32461160 | 0.12 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr14_+_35906366 | 0.11 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr23_+_31888903 | 0.11 |
ENSDART00000075730
|
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_+_22314304 | 0.11 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr19_+_43524098 | 0.11 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr10_+_37033 | 0.11 |
ENSDART00000157786
|
riox2
|
ribosomal oxygenase 2 |
| chr4_+_8007802 | 0.11 |
ENSDART00000014036
|
optn
|
optineurin |
| chr2_+_3202934 | 0.11 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr4_+_45204456 | 0.11 |
ENSDART00000158744
|
BX927336.3
|
ENSDARG00000102848 |
| chr23_+_17428538 | 0.11 |
|
|
|
| chr7_-_667670 | 0.11 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr16_-_11908084 | 0.11 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr16_+_29696109 | 0.10 |
ENSDART00000153683
|
VPS72
|
vacuolar protein sorting 72 homolog |
| chr13_+_36831946 | 0.10 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr12_+_26971596 | 0.10 |
|
|
|
| chr4_-_12915292 | 0.10 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.3 | 0.8 | GO:0019731 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.2 | 0.8 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.6 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.2 | 0.5 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.7 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.4 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.8 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.4 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 1.2 | GO:0035462 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.1 | 0.3 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.1 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.8 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.2 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.2 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.1 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.1 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.4 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.7 | GO:1903707 | negative regulation of hemopoiesis(GO:1903707) |
| 0.0 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.2 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.5 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 1.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.8 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.4 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.7 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.8 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |