Project
DANIO-CODE
Navigation
Downloads
Results for relb
Z-value: 0.66
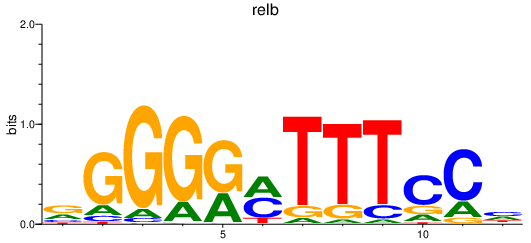
Transcription factors associated with relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
relb
|
ENSDARG00000086173 | v-rel avian reticuloendotheliosis viral oncogene homolog B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| relb | dr10_dc_chr15_+_17164373_17164429 | 0.23 | 4.0e-01 | Click! |
Activity profile of relb motif
Sorted Z-values of relb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of relb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_37418967 | 1.57 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr11_+_34788205 | 1.23 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr3_-_39316317 | 1.17 |
|
|
|
| chr8_+_39762186 | 1.14 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr2_+_21698100 | 1.09 |
ENSDART00000145670
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr3_-_22240424 | 1.07 |
|
|
|
| chr7_+_40878834 | 0.98 |
|
|
|
| chr16_+_35582277 | 0.83 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr7_-_37283707 | 0.81 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr6_-_10492123 | 0.76 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr1_-_30299017 | 0.66 |
|
|
|
| chr10_-_1933874 | 0.64 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_-_55211053 | 0.61 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr6_-_10492276 | 0.60 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr2_-_37419017 | 0.58 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr21_+_15660479 | 0.56 |
ENSDART00000101956
ENSDART00000026903 |
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr5_+_58564789 | 0.53 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr8_-_1669966 | 0.52 |
|
|
|
| chr13_+_36832090 | 0.49 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr23_-_33811895 | 0.49 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr19_-_7151715 | 0.47 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr1_-_54585975 | 0.47 |
ENSDART00000152504
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr5_+_58564645 | 0.47 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr23_-_18671263 | 0.46 |
ENSDART00000008847
|
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr15_+_6462447 | 0.46 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr19_+_46667326 | 0.43 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr10_-_1933761 | 0.42 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_-_30216377 | 0.42 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr3_-_9755642 | 0.42 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr5_-_11074111 | 0.41 |
|
|
|
| chr25_-_6263768 | 0.40 |
ENSDART00000166084
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr22_+_21996941 | 0.39 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr8_-_50899562 | 0.38 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long-chain family member 2 |
| chr3_-_3646681 | 0.38 |
ENSDART00000165648
ENSDART00000046454 ENSDART00000115282 ENSDART00000164463 |
ENSDARG00000076771
|
ENSDARG00000076771 |
| chr4_-_12782246 | 0.37 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr13_-_4863766 | 0.37 |
ENSDART00000159679
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr25_+_15901398 | 0.36 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr16_+_35582411 | 0.36 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr3_+_17728616 | 0.36 |
ENSDART00000167953
|
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr23_+_17428538 | 0.35 |
|
|
|
| chr20_+_16982314 | 0.35 |
ENSDART00000146625
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr2_-_37555425 | 0.35 |
ENSDART00000143496
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr5_+_57050210 | 0.34 |
|
|
|
| chr5_+_23582676 | 0.33 |
ENSDART00000139520
|
tp53
|
tumor protein p53 |
| chr1_+_54059634 | 0.32 |
ENSDART00000162075
|
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr22_+_1445227 | 0.31 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr16_-_24916921 | 0.31 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr11_+_36093297 | 0.30 |
ENSDART00000077649
|
sypl2a
|
synaptophysin-like 2a |
| chr21_+_21242470 | 0.30 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr13_+_22586873 | 0.29 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr5_-_67395562 | 0.29 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr2_+_21516908 | 0.28 |
ENSDART00000079518
|
dnajc1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr21_+_19381959 | 0.27 |
ENSDART00000080110
|
amacr
|
alpha-methylacyl-CoA racemase |
| chr5_+_23582805 | 0.27 |
ENSDART00000137298
|
tp53
|
tumor protein p53 |
| chr7_+_6629933 | 0.26 |
|
|
|
| chr3_-_39316355 | 0.26 |
|
|
|
| chr13_-_46424420 | 0.25 |
|
|
|
| chr24_+_5863770 | 0.25 |
ENSDART00000161104
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr23_-_18671336 | 0.25 |
ENSDART00000008847
|
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr22_-_2828572 | 0.24 |
|
|
|
| chr13_+_15685745 | 0.24 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr6_-_10492439 | 0.23 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr17_+_12788541 | 0.23 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr24_-_21788942 | 0.22 |
ENSDART00000113092
|
abhd10b
|
abhydrolase domain containing 10b |
| chr11_-_42934175 | 0.22 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr22_-_10410949 | 0.22 |
ENSDART00000111962
ENSDART00000064809 |
nol8
|
nucleolar protein 8 |
| chr2_-_30216703 | 0.22 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr15_+_38113499 | 0.21 |
|
|
|
| chr5_+_23582396 | 0.20 |
ENSDART00000051549
ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr7_+_13130367 | 0.20 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr7_-_50546498 | 0.20 |
|
|
|
| chr2_-_32755182 | 0.19 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr20_-_5008341 | 0.19 |
|
|
|
| chr15_+_17164373 | 0.18 |
ENSDART00000123197
ENSDART00000154418 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr23_+_3570243 | 0.18 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr25_-_34634739 | 0.18 |
ENSDART00000157370
|
CR762436.3
|
Histone H3.2 |
| chr13_+_36831946 | 0.18 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr9_-_9264634 | 0.17 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr21_+_13689239 | 0.17 |
ENSDART00000112451
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr25_-_10532989 | 0.16 |
ENSDART00000177834
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr11_+_40280114 | 0.16 |
ENSDART00000175424
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr25_-_6263829 | 0.16 |
ENSDART00000166084
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr12_+_23939657 | 0.16 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr13_-_34667689 | 0.16 |
ENSDART00000025719
|
ism1
|
isthmin 1 |
| chr15_+_38113014 | 0.15 |
|
|
|
| chr3_-_31487744 | 0.15 |
ENSDART00000124559
|
moto
|
minamoto |
| chr5_+_6391432 | 0.15 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr10_+_42714071 | 0.15 |
ENSDART00000005496
|
kctd9b
|
potassium channel tetramerization domain containing 9b |
| chr15_-_23594718 | 0.14 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr25_+_34546629 | 0.14 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr25_+_15901561 | 0.14 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr6_+_3174047 | 0.14 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr13_+_22589282 | 0.14 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_-_30216240 | 0.13 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr16_-_32174276 | 0.13 |
ENSDART00000137699
|
cnot3a
|
CCR4-NOT transcription complex, subunit 3a |
| chr20_-_21775455 | 0.13 |
|
|
|
| chr18_-_16934961 | 0.13 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr15_+_6462607 | 0.13 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr5_+_24378828 | 0.13 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr24_-_11365575 | 0.12 |
ENSDART00000140217
|
prpf4bb
|
pre-mRNA processing factor 4Bb |
| chr15_-_37202518 | 0.12 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr2_-_30216333 | 0.12 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr8_-_17755104 | 0.12 |
ENSDART00000131276
|
si:ch211-150o23.2
|
si:ch211-150o23.2 |
| chr5_-_43219303 | 0.12 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr5_-_28522443 | 0.12 |
ENSDART00000144802
|
dfnb31b
|
deafness, autosomal recessive 31b |
| chr21_+_19511167 | 0.11 |
ENSDART00000058487
|
rai14
|
retinoic acid induced 14 |
| chr15_+_17164535 | 0.11 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr21_+_21754830 | 0.10 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr2_+_8981028 | 0.10 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr9_-_11291927 | 0.10 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr20_+_31375056 | 0.10 |
|
|
|
| chr9_+_42805317 | 0.10 |
ENSDART00000138133
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr7_+_40879017 | 0.10 |
|
|
|
| chr1_-_22603603 | 0.08 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr20_+_31375136 | 0.08 |
|
|
|
| chr15_-_38113404 | 0.08 |
|
|
|
| chr9_+_492726 | 0.08 |
ENSDART00000112635
|
ENSDARG00000078172
|
ENSDARG00000078172 |
| chr19_-_17039269 | 0.07 |
|
|
|
| chr25_+_34557335 | 0.07 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr7_+_13130332 | 0.07 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr7_-_37283894 | 0.06 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr12_-_28732946 | 0.04 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr19_-_307085 | 0.04 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr5_-_52318158 | 0.04 |
ENSDART00000166267
|
zfpl1
|
zinc finger protein-like 1 |
| chr3_+_35276575 | 0.04 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr1_+_28407560 | 0.04 |
ENSDART00000053924
|
irs2a
|
insulin receptor substrate 2a |
| chr20_+_23602537 | 0.03 |
|
|
|
| chr25_+_15901662 | 0.03 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr8_-_12394879 | 0.03 |
ENSDART00000101174
|
traf1
|
TNF receptor-associated factor 1 |
| chr17_-_28794581 | 0.03 |
ENSDART00000132811
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr17_+_51593393 | 0.03 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr13_+_36832145 | 0.02 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr14_-_6096258 | 0.02 |
|
|
|
| chr20_-_6153384 | 0.02 |
ENSDART00000161566
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr22_-_37092134 | 0.02 |
|
|
|
| chr6_-_48474815 | 0.01 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr23_-_36349563 | 0.00 |
|
|
|
| chr13_-_22964527 | 0.00 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.4 | 1.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.2 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 0.6 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.8 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 0.3 | GO:0098920 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0010939 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:1903707 | negative regulation of hemopoiesis(GO:1903707) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |