Project
DANIO-CODE
Navigation
Downloads
Results for rest
Z-value: 3.70
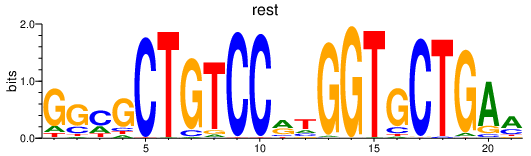
Transcription factors associated with rest
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rest
|
ENSDARG00000103046 | Danio rerio RE1-silencing transcription factor (rest), mRNA. |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rest | dr10_dc_chr14_-_51717514_51717624 | -0.14 | 6.0e-01 | Click! |
Activity profile of rest motif
Sorted Z-values of rest motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rest
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_30858769 | 25.09 |
ENSDART00000131285
|
myt1a
|
myelin transcription factor 1a |
| chr20_+_35010140 | 14.23 |
ENSDART00000172001
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr12_+_24221087 | 13.61 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr1_+_16983775 | 12.98 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr1_+_10036098 | 12.55 |
ENSDART00000139387
|
atp1b1b
|
ATPase, Na+/K+ transporting, beta 1b polypeptide |
| chr4_-_5294280 | 11.82 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr25_-_22541525 | 11.73 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_+_28252623 | 11.49 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr21_+_6829161 | 10.27 |
ENSDART00000037265
|
olfm1b
|
olfactomedin 1b |
| chr25_-_31452381 | 10.02 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr10_+_21850059 | 9.79 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr9_-_35106146 | 9.53 |
ENSDART00000139608
ENSDART00000100728 ENSDART00000123005 |
upf3a
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr13_+_781725 | 8.48 |
ENSDART00000129866
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr13_+_23152038 | 8.44 |
ENSDART00000171676
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr6_+_41257803 | 8.35 |
ENSDART00000042683
|
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr18_+_36825511 | 7.97 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr14_-_2274883 | 7.95 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr2_+_57369722 | 7.89 |
ENSDART00000129811
|
onecut3b
|
one cut homeobox 3b |
| chr23_-_45356039 | 7.75 |
ENSDART00000067630
|
ENSDARG00000039901
|
ENSDARG00000039901 |
| chr7_+_23636549 | 7.69 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr5_-_25493792 | 7.66 |
ENSDART00000010199
|
fam219ab
|
family with sequence similarity 219, member Ab |
| chr7_+_31608828 | 7.63 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr25_+_34745872 | 7.59 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr25_+_4909062 | 7.44 |
ENSDART00000170049
|
parvb
|
parvin, beta |
| chr20_-_54669179 | 7.43 |
|
|
|
| chr2_-_28154388 | 7.26 |
ENSDART00000161864
|
zgc:123035
|
zgc:123035 |
| chr23_+_35660991 | 7.19 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr15_+_24277664 | 6.89 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr3_+_33209227 | 6.82 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr7_-_32562435 | 6.78 |
ENSDART00000099872
ENSDART00000099871 ENSDART00000147554 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr10_+_23052796 | 6.77 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr9_-_27838603 | 6.70 |
|
|
|
| chr17_-_12231165 | 6.49 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr12_-_1931281 | 6.16 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr9_-_30073641 | 6.06 |
|
|
|
| chr6_-_42143595 | 6.06 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr20_+_38821682 | 6.02 |
ENSDART00000015095
|
uts1
|
urotensin 1 |
| chr6_-_43094573 | 5.95 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr5_+_43365449 | 5.93 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr23_+_36648211 | 5.84 |
|
|
|
| chr14_-_31918174 | 5.72 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr4_-_5293599 | 5.56 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr10_+_21630540 | 5.53 |
|
|
|
| chr3_+_56008829 | 5.52 |
|
|
|
| chr14_-_2155141 | 5.51 |
ENSDART00000106691
|
pcdh2g6
|
protocadherin 2 gamma 6 |
| chr21_+_11375961 | 5.45 |
ENSDART00000158936
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr11_+_35896325 | 5.44 |
ENSDART00000163330
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr14_-_2175076 | 5.36 |
ENSDART00000110937
|
CT573264.3
|
ENSDARG00000079424 |
| chr14_-_2179471 | 5.29 |
ENSDART00000157401
|
pcdh2g1
|
protocadherin 2 gamma 1 |
| chr7_+_31608878 | 5.17 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr2_+_56705456 | 5.05 |
|
|
|
| chr19_-_4769990 | 5.00 |
|
|
|
| chr5_+_71028018 | 4.98 |
ENSDART00000164893
ENSDART00000159658 ENSDART00000097164 ENSDART00000124939 ENSDART00000171230 |
LHX3
|
LIM homeobox 3 |
| chr7_+_39957385 | 4.97 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr5_-_29150278 | 4.72 |
ENSDART00000051474
|
barhl1a
|
BarH-like homeobox 1a |
| chr11_-_25962291 | 4.61 |
|
|
|
| chr19_-_42674928 | 4.59 |
ENSDART00000102697
|
cart4
|
cocaine- and amphetamine-regulated transcript 4 |
| chr20_+_5511996 | 4.53 |
|
|
|
| chr19_+_27133834 | 4.36 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr1_+_33510660 | 4.27 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr6_+_10214270 | 4.25 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr18_+_29010470 | 4.17 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr20_+_27121126 | 4.17 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr2_-_5864098 | 3.83 |
ENSDART00000133851
|
nyap2b
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2b |
| chr13_+_23152160 | 3.80 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr21_+_6829352 | 3.74 |
ENSDART00000146371
|
olfm1b
|
olfactomedin 1b |
| chr3_-_59179368 | 3.67 |
ENSDART00000100327
|
nptx1l
|
neuronal pentraxin 1 like |
| chr3_-_26073923 | 3.62 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr15_+_43535893 | 3.57 |
|
|
|
| chr15_-_22212082 | 3.57 |
ENSDART00000159116
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr3_-_38603604 | 3.52 |
ENSDART00000165823
|
si:ch211-198m17.1
|
si:ch211-198m17.1 |
| chr14_-_2159010 | 3.30 |
ENSDART00000158928
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr20_+_45837841 | 2.97 |
ENSDART00000113454
|
chgb
|
chromogranin B |
| chr17_-_23235070 | 2.96 |
|
|
|
| chr14_-_2162867 | 2.76 |
ENSDART00000159262
|
pcdh2g3
|
protocadherin 2 gamma 3 |
| chr5_+_43365528 | 2.59 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr11_+_19618349 | 2.56 |
ENSDART00000157394
|
cadpsa
|
Ca2+-dependent activator protein for secretion a |
| chr6_+_19680394 | 2.54 |
|
|
|
| chr7_+_16978182 | 2.53 |
|
|
|
| chr7_+_16977286 | 2.45 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr14_-_2349346 | 2.13 |
ENSDART00000130712
|
pcdh2aa15
|
protocadherin 2 alpha a 15 |
| chr7_+_16977507 | 2.04 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr9_-_51251505 | 1.81 |
ENSDART00000162278
|
CABZ01053748.1
|
ENSDARG00000100870 |
| chr14_-_2114123 | 1.60 |
ENSDART00000111335
|
pcdh2g13
|
protocadherin 2 gamma 13 |
| chr6_+_43905694 | 1.54 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr20_-_31028201 | 1.48 |
ENSDART00000146376
|
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr14_-_2166955 | 1.47 |
ENSDART00000169201
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr4_+_344355 | 1.46 |
ENSDART00000142225
ENSDART00000163436 |
tmem181
|
transmembrane protein 181 |
| chr11_+_39631586 | 1.43 |
ENSDART00000173280
|
camta1b
|
calmodulin binding transcription activator 1b |
| chr23_+_20714105 | 1.07 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr10_-_30006277 | 1.05 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr10_+_21609883 | 0.96 |
|
|
|
| chr17_+_19610407 | 0.84 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr3_+_34690823 | 0.83 |
|
|
|
| chr1_+_54233842 | 0.82 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr18_+_29010525 | 0.74 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr3_-_13771322 | 0.73 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr21_-_17446442 | 0.45 |
|
|
|
| chr3_-_26073676 | 0.38 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr18_-_44617805 | 0.30 |
ENSDART00000173095
ENSDART00000136420 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_+_19183576 | 0.22 |
ENSDART00000066391
|
csnk1e
|
casein kinase 1, epsilon |
| chr25_-_31452269 | 0.20 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr10_-_26775840 | 0.14 |
ENSDART00000020096
|
fgf13b
|
fibroblast growth factor 13b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 13.0 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 3.2 | 12.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 2.9 | 14.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 2.2 | 10.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.7 | 20.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 1.6 | 11.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.2 | 3.6 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 1.2 | 5.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.1 | 4.5 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 1.0 | 4.2 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 1.0 | 11.5 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.9 | 5.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.9 | 6.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.9 | 4.3 | GO:0008291 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.9 | 6.0 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 12.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.8 | 8.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.7 | 12.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.5 | 6.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.5 | 10.2 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.3 | 39.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 7.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.3 | 9.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 1.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.2 | 11.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.2 | 12.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 13.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 25.1 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.1 | 0.3 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 4.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 3.5 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 2.8 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 1.5 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 1.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 5.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 4.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.8 | 5.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.6 | 12.5 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 1.5 | 20.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 1.4 | 14.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 10.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.7 | 4.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 3.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.6 | 10.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 4.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 5.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 7.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 7.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 40.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 8.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 13.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 14.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 6.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 33.0 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 6.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 12.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 44.0 | GO:0016021 | integral component of membrane(GO:0016021) intrinsic component of membrane(GO:0031224) |
| 0.0 | 53.8 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 3.6 | 14.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 3.2 | 13.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 1.6 | 12.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.4 | 5.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.0 | 13.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.9 | 20.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.9 | 3.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.9 | 4.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.8 | 5.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 7.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.7 | 12.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.7 | 12.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.5 | 12.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.5 | 1.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.4 | 4.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 9.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 6.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 4.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 39.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 25.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 6.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 13.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 7.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.4 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 1.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 24.3 | GO:0046872 | metal ion binding(GO:0046872) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 7.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 4.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 8.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 13.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 9.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |