Project
DANIO-CODE
Navigation
Downloads
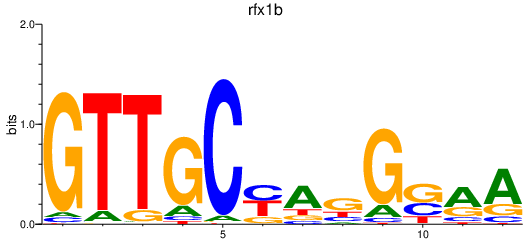
Results for rfx1b
Z-value: 0.44
Transcription factors associated with rfx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx1b
|
ENSDARG00000075904 | regulatory factor X, 1b (influences HLA class II expression) |
Activity profile of rfx1b motif
Sorted Z-values of rfx1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_33527870 | 0.86 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr17_-_10682357 | 0.82 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr1_+_21309690 | 0.68 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr16_-_9978112 | 0.67 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr12_+_21570808 | 0.56 |
|
|
|
| chr9_+_9133904 | 0.56 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr25_+_25358457 | 0.55 |
ENSDART00000112330
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr3_-_59884282 | 0.54 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr3_+_49277125 | 0.53 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr17_-_29102320 | 0.50 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr23_+_10534968 | 0.48 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr13_-_47266563 | 0.47 |
ENSDART00000131239
|
acoxl
|
acyl-CoA oxidase-like |
| chr2_-_43110857 | 0.47 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr14_+_23986703 | 0.46 |
ENSDART00000079164
|
klhl3
|
kelch-like family member 3 |
| chr5_+_61256474 | 0.46 |
ENSDART00000097340
|
rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr11_+_7518953 | 0.44 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr22_-_36721626 | 0.44 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr7_-_33921565 | 0.43 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr2_+_33352680 | 0.41 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_+_30894667 | 0.39 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr1_-_48853800 | 0.38 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr3_+_45375639 | 0.37 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr21_+_43674754 | 0.37 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr9_+_48717490 | 0.37 |
ENSDART00000163353
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr6_-_39661423 | 0.36 |
ENSDART00000110380
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr16_+_9141325 | 0.35 |
|
|
|
| chr14_+_6647838 | 0.35 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr16_-_16682692 | 0.35 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr1_-_44942144 | 0.34 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr21_-_1600125 | 0.34 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr11_+_44460956 | 0.34 |
ENSDART00000168531
|
irf2bp2b
|
interferon regulatory factor 2 binding protein 2b |
| chr8_-_49356479 | 0.34 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr9_+_41072345 | 0.32 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr10_+_35344246 | 0.32 |
ENSDART00000139229
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr25_+_3884499 | 0.31 |
ENSDART00000104926
|
ENSDARG00000058108
|
ENSDARG00000058108 |
| chr17_+_21944679 | 0.30 |
ENSDART00000063704
|
crip3
|
cysteine-rich protein 3 |
| chr17_+_34238628 | 0.30 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr6_-_8029184 | 0.30 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr13_-_51662834 | 0.30 |
|
|
|
| chr15_-_45229713 | 0.29 |
|
|
|
| chr3_+_27684309 | 0.28 |
|
|
|
| chr24_-_475010 | 0.28 |
ENSDART00000108762
|
vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr24_-_38758093 | 0.28 |
|
|
|
| chr21_+_9483423 | 0.27 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr21_-_43611307 | 0.27 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr3_-_12087518 | 0.27 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr1_-_44942047 | 0.27 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr8_+_7340538 | 0.27 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr3_-_42649955 | 0.26 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr13_-_7435355 | 0.26 |
ENSDART00000121952
|
h2afy2
|
H2A histone family, member Y2 |
| chr24_+_38267919 | 0.26 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr20_-_3970778 | 0.26 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr19_-_40599163 | 0.26 |
ENSDART00000150972
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr16_+_41922320 | 0.26 |
|
|
|
| chr17_+_43605118 | 0.26 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr2_+_6214792 | 0.26 |
ENSDART00000153916
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr5_+_38963156 | 0.26 |
ENSDART00000097872
|
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr17_+_8831186 | 0.25 |
ENSDART00000109573
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr24_-_16922912 | 0.25 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr9_+_48521741 | 0.25 |
ENSDART00000145972
|
ccdc173
|
coiled-coil domain containing 173 |
| chr11_-_37730181 | 0.24 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr9_-_22306541 | 0.24 |
ENSDART00000101916
|
crygm2d15
|
crystallin, gamma M2d15 |
| chr15_-_23271279 | 0.24 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr19_-_5142310 | 0.23 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr7_+_26523177 | 0.23 |
|
|
|
| chr13_-_31339870 | 0.23 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr24_-_37311408 | 0.23 |
ENSDART00000137777
|
dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr3_+_45375698 | 0.23 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr17_-_14551031 | 0.23 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr4_-_18950323 | 0.23 |
ENSDART00000132081
ENSDART00000042250 |
rap1b
|
RAP1B, member of RAS oncogene family |
| chr6_+_44891187 | 0.23 |
|
|
|
| chr11_-_30105047 | 0.23 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr2_+_294913 | 0.22 |
ENSDART00000130999
|
zgc:113452
|
zgc:113452 |
| chr16_+_5302404 | 0.22 |
ENSDART00000145368
|
soga3a
|
SOGA family member 3a |
| chr7_-_34234508 | 0.22 |
ENSDART00000173806
|
tex9
|
testis expressed 9 |
| chr4_-_72087221 | 0.22 |
|
|
|
| chr19_-_34328673 | 0.21 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr19_-_46396376 | 0.21 |
|
|
|
| chr15_+_28435937 | 0.21 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr5_+_69096325 | 0.20 |
ENSDART00000014649
|
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr6_-_26897413 | 0.20 |
|
|
|
| chr23_+_21453261 | 0.20 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr16_-_7321943 | 0.19 |
ENSDART00000149260
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr24_+_42884 | 0.19 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr24_-_28576494 | 0.18 |
|
|
|
| chr8_+_7937320 | 0.18 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr20_+_36779159 | 0.18 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr17_-_24666272 | 0.18 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr14_-_3264798 | 0.18 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr10_-_15896595 | 0.17 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr10_-_36738730 | 0.17 |
ENSDART00000139097
|
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr5_+_29071526 | 0.17 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr6_+_21779793 | 0.17 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr5_+_36487425 | 0.17 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr8_+_5222065 | 0.16 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr22_+_24686610 | 0.16 |
ENSDART00000088027
|
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr2_-_45810868 | 0.16 |
ENSDART00000135665
ENSDART00000075080 |
prpf38b
|
pre-mRNA processing factor 38B |
| chr14_+_23980348 | 0.16 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr16_-_31835463 | 0.16 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr6_+_3673621 | 0.15 |
ENSDART00000138185
|
gad1b
|
glutamate decarboxylase 1b |
| chr4_+_19545842 | 0.15 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr17_+_23975487 | 0.15 |
|
|
|
| chr20_-_54669179 | 0.15 |
|
|
|
| chr10_-_29858525 | 0.15 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_+_49894978 | 0.15 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr13_+_32987105 | 0.15 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr20_+_31173383 | 0.15 |
ENSDART00000136255
|
otofa
|
otoferlin a |
| chr17_+_27157965 | 0.15 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr20_-_28739099 | 0.15 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr16_+_24047649 | 0.14 |
|
|
|
| chr18_-_31073514 | 0.14 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr7_-_54407681 | 0.14 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr20_-_4723724 | 0.14 |
ENSDART00000147071
|
ak7a
|
adenylate kinase 7a |
| chr10_-_8029671 | 0.14 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr15_+_14048904 | 0.14 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr20_+_9776103 | 0.14 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr1_-_20845715 | 0.14 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr17_-_29102271 | 0.14 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr8_-_50159285 | 0.13 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr17_-_31678247 | 0.13 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr19_-_28382815 | 0.13 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr13_-_18559962 | 0.13 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr4_-_20593689 | 0.13 |
ENSDART00000066894
|
dcp1b
|
decapping mRNA 1B |
| chr12_+_17498203 | 0.13 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr16_+_42161483 | 0.13 |
|
|
|
| chr2_-_42784707 | 0.13 |
|
|
|
| chr21_-_485916 | 0.13 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr13_+_12474311 | 0.13 |
ENSDART00000145136
|
metap1
|
methionyl aminopeptidase 1 |
| chr22_-_12723704 | 0.13 |
ENSDART00000080741
|
plcd4a
|
phospholipase C, delta 4a |
| chr7_-_40467922 | 0.13 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr8_-_46037132 | 0.13 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr20_-_38714872 | 0.12 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| KN150503v1_-_598 | 0.12 |
ENSDART00000174608
|
CABZ01110189.1
|
ENSDARG00000108333 |
| chr1_-_5527494 | 0.12 |
ENSDART00000131254
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
| chr24_+_192889 | 0.12 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr13_-_31340029 | 0.12 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr4_+_15820977 | 0.12 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr17_-_8016214 | 0.12 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr12_+_33261056 | 0.12 |
ENSDART00000124982
|
ccdc57
|
coiled-coil domain containing 57 |
| chr16_+_29697357 | 0.12 |
|
|
|
| chr15_-_10345337 | 0.12 |
|
|
|
| chr1_+_47140627 | 0.12 |
ENSDART00000164073
ENSDART00000101066 ENSDART00000110335 |
cfap58
|
cilia and flagella associated protein 58 |
| chr8_-_30233570 | 0.12 |
ENSDART00000143809
|
zgc:162939
|
zgc:162939 |
| chr3_+_60549501 | 0.12 |
|
|
|
| chr17_-_22047445 | 0.12 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr19_+_17941678 | 0.12 |
ENSDART00000124597
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr2_-_2456819 | 0.11 |
ENSDART00000013794
ENSDART00000082155 ENSDART00000169871 ENSDART00000167202 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr8_-_22252973 | 0.11 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr3_-_19050721 | 0.11 |
ENSDART00000131503
|
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr2_+_7203572 | 0.11 |
ENSDART00000022963
|
cdc14aa
|
cell division cycle 14Aa |
| chr25_-_10532989 | 0.11 |
ENSDART00000177834
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr24_+_36516320 | 0.11 |
|
|
|
| chr24_-_9819862 | 0.11 |
ENSDART00000092975
|
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr12_-_28840796 | 0.11 |
|
|
|
| chr7_+_26788074 | 0.11 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr11_-_18637950 | 0.10 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr13_+_24587429 | 0.10 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr1_+_49894685 | 0.10 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr18_+_29971911 | 0.10 |
ENSDART00000146431
ENSDART00000099285 |
atmin
|
ATM interactor |
| chr11_+_7518748 | 0.10 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr13_+_28723589 | 0.10 |
|
|
|
| chr21_-_36495488 | 0.10 |
|
|
|
| chr2_+_57995336 | 0.10 |
|
|
|
| chr14_+_35552512 | 0.09 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr1_+_52645062 | 0.09 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr15_-_37687938 | 0.09 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr8_+_2428689 | 0.09 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr25_-_20426215 | 0.09 |
|
|
|
| chr6_+_60190849 | 0.09 |
|
|
|
| chr17_-_33764165 | 0.09 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr18_-_17542397 | 0.08 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr18_-_43890514 | 0.08 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr24_-_39884167 | 0.08 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr22_+_1360497 | 0.08 |
ENSDART00000160330
|
BX322657.1
|
ENSDARG00000102448 |
| chr1_-_46027053 | 0.08 |
ENSDART00000074514
|
spryd7a
|
SPRY domain containing 7a |
| chr23_+_24662595 | 0.08 |
ENSDART00000078805
|
mlh3
|
mutL homolog 3 (E. coli) |
| chr17_+_11346554 | 0.08 |
ENSDART00000113669
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr4_+_32755927 | 0.08 |
|
|
|
| chr10_-_17274395 | 0.08 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr4_-_18617553 | 0.08 |
ENSDART00000171450
ENSDART00000172976 |
CDPF1
|
cysteine rich DPF motif domain containing 1 |
| chr6_-_10681098 | 0.08 |
|
|
|
| chr16_+_41767229 | 0.08 |
ENSDART00000075937
|
mtdhb
|
metadherin b |
| chr18_-_44365869 | 0.08 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr13_-_48465147 | 0.07 |
ENSDART00000132895
|
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr4_-_1952201 | 0.07 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr4_+_13651618 | 0.07 |
ENSDART00000110370
|
nrcama
|
neuronal cell adhesion molecule a |
| chr5_+_36049885 | 0.07 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr3_+_45375580 | 0.07 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr5_-_22847399 | 0.07 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr12_-_196397 | 0.07 |
ENSDART00000161178
ENSDART00000160926 |
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr15_+_32863182 | 0.07 |
|
|
|
| chr25_+_35437993 | 0.06 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_+_24551284 | 0.06 |
ENSDART00000088238
|
slc25a17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein), member 17 |
| chr4_-_18660286 | 0.06 |
ENSDART00000178638
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr25_+_25343112 | 0.06 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr14_-_46646301 | 0.06 |
|
|
|
| chr11_-_3592968 | 0.06 |
ENSDART00000085855
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr20_+_27104580 | 0.06 |
ENSDART00000152956
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr5_-_66145078 | 0.06 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr18_+_16203004 | 0.06 |
ENSDART00000135016
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 0.7 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.2 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.8 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.1 | 0.2 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.1 | 0.5 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.2 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.2 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.5 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.3 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |