Project
DANIO-CODE
Navigation
Downloads
Results for rfx3_rfx1a_rfx2
Z-value: 2.45
Transcription factors associated with rfx3_rfx1a_rfx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
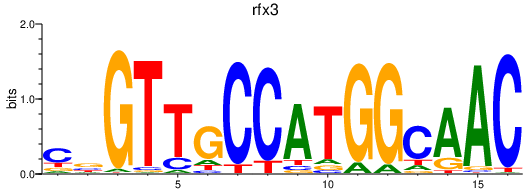
rfx3
|
ENSDARG00000014550 | regulatory factor X, 3 (influences HLA class II expression) |
|
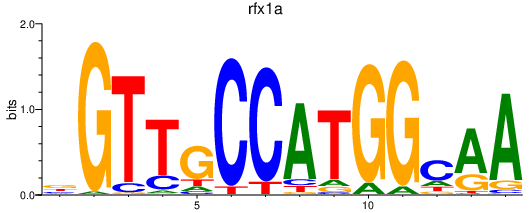
rfx1a
|
ENSDARG00000005883 | regulatory factor X, 1a (influences HLA class II expression) |
|
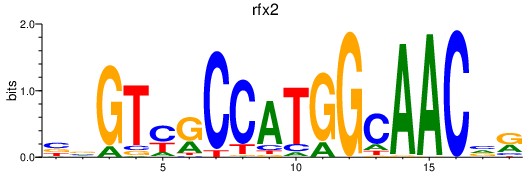
rfx2
|
ENSDARG00000013575 | regulatory factor X, 2 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx2 | dr10_dc_chr8_-_20106132_20106149 | -0.86 | 2.1e-05 | Click! |
| rfx1a | dr10_dc_chr3_-_19050721_19050795 | 0.70 | 2.4e-03 | Click! |
| rfx3 | dr10_dc_chr10_-_625501_625665 | 0.20 | 4.6e-01 | Click! |
Activity profile of rfx3_rfx1a_rfx2 motif
Sorted Z-values of rfx3_rfx1a_rfx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx3_rfx1a_rfx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_475010 | 15.93 |
ENSDART00000108762
|
vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr16_-_9978112 | 7.96 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr8_+_21163236 | 7.55 |
ENSDART00000091307
|
col2a1a
|
collagen, type II, alpha 1a |
| chr2_+_57995336 | 7.10 |
|
|
|
| chr5_-_70948223 | 7.05 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr23_+_35609887 | 6.86 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr13_-_13163676 | 6.48 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr16_-_45103102 | 5.88 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr2_-_30675594 | 5.25 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr7_-_57796486 | 5.20 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr25_+_25358457 | 4.52 |
ENSDART00000112330
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr22_+_16511562 | 4.49 |
|
|
|
| chr11_+_6118409 | 4.30 |
ENSDART00000027666
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr9_+_53736715 | 4.26 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr21_-_15833030 | 4.23 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr3_+_60637167 | 4.16 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr8_-_49356479 | 4.12 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr17_-_10682357 | 4.09 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr14_-_25279835 | 3.95 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr6_-_8501230 | 3.87 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr21_+_25034997 | 3.80 |
ENSDART00000167523
|
dixdc1b
|
DIX domain containing 1b |
| chr21_-_1600125 | 3.70 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr15_+_47316316 | 3.69 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr13_-_13163801 | 3.61 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr21_-_43806271 | 3.56 |
ENSDART00000138710
|
pdlim4
|
PDZ and LIM domain 4 |
| chr8_+_2428689 | 3.53 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr6_+_13833991 | 3.52 |
|
|
|
| chr10_+_5693933 | 3.50 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_26872047 | 3.42 |
ENSDART00000138693
|
si:ch211-106k21.5
|
si:ch211-106k21.5 |
| chr1_-_10122492 | 3.31 |
ENSDART00000139749
|
ENSDARG00000087107
|
ENSDARG00000087107 |
| chr25_+_36878565 | 3.29 |
ENSDART00000157596
|
ric3b
|
RIC3 acetylcholine receptor chaperone b |
| chr25_-_2485276 | 3.17 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr12_-_36191048 | 3.15 |
ENSDART00000177986
|
cep131
|
centrosomal protein 131 |
| chr9_-_56292487 | 3.11 |
ENSDART00000151720
|
si:ch211-39i22.1
|
si:ch211-39i22.1 |
| chr13_-_31339870 | 3.11 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr2_-_43110857 | 3.08 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr22_+_2921305 | 3.05 |
ENSDART00000143258
|
cep19
|
centrosomal protein 19 |
| chr21_+_26660833 | 3.02 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr19_-_5142310 | 2.99 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr9_-_16845777 | 2.98 |
ENSDART00000160273
|
ENSDARG00000102261
|
ENSDARG00000102261 |
| chr17_+_8831186 | 2.97 |
ENSDART00000109573
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr16_-_29593569 | 2.95 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr4_-_376986 | 2.89 |
ENSDART00000067482
ENSDART00000160718 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr14_+_4489377 | 2.89 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_35745559 | 2.85 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr8_+_2260519 | 2.80 |
ENSDART00000136743
|
ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr7_-_28278286 | 2.79 |
ENSDART00000113313
|
st5
|
suppression of tumorigenicity 5 |
| chr9_-_6949104 | 2.76 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr1_-_55554518 | 2.76 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr16_+_49796978 | 2.71 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr5_-_22675495 | 2.70 |
ENSDART00000161883
|
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr7_+_49628680 | 2.66 |
ENSDART00000007487
|
apip
|
APAF1 interacting protein |
| chr24_-_38928988 | 2.62 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr8_+_7740132 | 2.62 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr21_+_11410738 | 2.58 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr9_-_44487895 | 2.56 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr15_-_45229713 | 2.52 |
|
|
|
| KN150001v1_+_19561 | 2.52 |
|
|
|
| chr6_+_52212513 | 2.50 |
ENSDART00000025940
|
ywhaba
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a |
| chr15_+_1187249 | 2.50 |
ENSDART00000152638
ENSDART00000152466 |
mlf1
|
myeloid leukemia factor 1 |
| chr12_+_25509394 | 2.50 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr5_+_38963156 | 2.48 |
ENSDART00000097872
|
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr24_+_17125429 | 2.48 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr3_+_54489642 | 2.44 |
ENSDART00000114443
|
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr3_-_12087518 | 2.44 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr8_-_34077790 | 2.43 |
|
|
|
| chr16_-_24697750 | 2.40 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr19_-_28382815 | 2.38 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr3_-_49242655 | 2.37 |
ENSDART00000142200
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr10_+_22759607 | 2.36 |
|
|
|
| chr19_-_40599163 | 2.35 |
ENSDART00000150972
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr2_+_6214792 | 2.35 |
ENSDART00000153916
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr21_+_22386643 | 2.34 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr16_+_10974655 | 2.33 |
|
|
|
| chr15_-_18226612 | 2.32 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr13_+_1051015 | 2.31 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr24_-_2918801 | 2.31 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr4_+_12618485 | 2.30 |
ENSDART00000112860
|
lmo3
|
LIM domain only 3 |
| chr7_+_40183670 | 2.27 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr17_+_52024610 | 2.27 |
|
|
|
| chr20_+_4120302 | 2.26 |
ENSDART00000159837
|
cnksr3
|
cnksr family member 3 |
| chr22_+_38028979 | 2.24 |
ENSDART00000165078
ENSDART00000159906 |
CU659306.1
|
ENSDARG00000100625 |
| chr16_-_31835463 | 2.24 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr1_-_44942144 | 2.23 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr25_+_32114076 | 2.19 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr25_-_25641897 | 2.18 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr24_-_38215159 | 2.15 |
|
|
|
| chr3_-_34208423 | 2.14 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr13_+_33115704 | 2.11 |
ENSDART00000141912
|
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr21_-_21477423 | 2.10 |
ENSDART00000164076
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr5_-_57250212 | 2.09 |
ENSDART00000023262
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr2_-_56224858 | 2.07 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr21_+_27427785 | 2.07 |
ENSDART00000113061
|
bbs1
|
Bardet-Biedl syndrome 1 |
| chr17_-_24666272 | 2.05 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr2_+_294913 | 2.05 |
ENSDART00000130999
|
zgc:113452
|
zgc:113452 |
| chr16_+_24717056 | 2.04 |
ENSDART00000157237
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr16_-_33976031 | 2.02 |
ENSDART00000110743
|
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr11_+_6118322 | 2.02 |
ENSDART00000165031
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr3_+_49277125 | 1.99 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr23_+_21453261 | 1.99 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr3_+_31813888 | 1.98 |
ENSDART00000114957
|
zgc:193811
|
zgc:193811 |
| chr16_+_9141325 | 1.98 |
|
|
|
| chr5_-_54026450 | 1.96 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr13_-_252864 | 1.93 |
|
|
|
| chr8_-_15267816 | 1.93 |
ENSDART00000137923
|
spata6
|
spermatogenesis associated 6 |
| chr15_-_44029439 | 1.91 |
|
|
|
| chr8_+_26502828 | 1.90 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr14_+_23220869 | 1.89 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr19_-_10324632 | 1.87 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr19_-_34328673 | 1.87 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr18_+_35198982 | 1.86 |
ENSDART00000127379
|
cfap45
|
cilia and flagella associated protein 45 |
| chr10_-_43200499 | 1.85 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr6_-_33039029 | 1.84 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr7_+_9726412 | 1.83 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr10_-_36738730 | 1.83 |
ENSDART00000139097
|
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr20_+_32646617 | 1.82 |
|
|
|
| chr25_+_13635079 | 1.82 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr21_-_22520832 | 1.81 |
ENSDART00000167230
|
cfap53
|
cilia and flagella associated protein 53 |
| chr1_-_44942047 | 1.81 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr13_-_36577270 | 1.80 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr21_-_22672597 | 1.80 |
ENSDART00000140032
ENSDART00000136993 |
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr2_+_47864913 | 1.80 |
|
|
|
| chr23_+_23559246 | 1.79 |
ENSDART00000172214
|
agrn
|
agrin |
| chr6_+_8956398 | 1.79 |
ENSDART00000150916
|
ENSDARG00000071159
|
ENSDARG00000071159 |
| chr2_-_42784609 | 1.77 |
|
|
|
| chr12_+_33261056 | 1.76 |
ENSDART00000124982
|
ccdc57
|
coiled-coil domain containing 57 |
| chr6_-_29297541 | 1.76 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr13_+_32987105 | 1.73 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr9_-_3700395 | 1.73 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr8_+_54058732 | 1.72 |
|
|
|
| chr21_+_142265 | 1.71 |
|
|
|
| chr23_+_22350713 | 1.71 |
ENSDART00000130375
|
rap1gap
|
RAP1 GTPase activating protein |
| chr6_+_47425082 | 1.69 |
ENSDART00000171087
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr23_-_26150495 | 1.68 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr5_-_54954988 | 1.68 |
ENSDART00000142912
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr6_-_2089453 | 1.67 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr4_-_5643440 | 1.67 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr19_-_9553977 | 1.67 |
ENSDART00000045565
|
vamp1
|
vesicle-associated membrane protein 1 |
| chr10_-_17274395 | 1.66 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr15_-_21003820 | 1.66 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr7_+_890752 | 1.65 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr18_-_39491932 | 1.62 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr1_+_54231447 | 1.61 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr10_+_31336284 | 1.61 |
ENSDART00000140988
|
tmem218
|
transmembrane protein 218 |
| chr11_+_36215554 | 1.61 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| KN150503v1_-_598 | 1.60 |
ENSDART00000174608
|
CABZ01110189.1
|
ENSDARG00000108333 |
| chr7_-_33994315 | 1.60 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr19_+_46647893 | 1.59 |
ENSDART00000162785
ENSDART00000171251 ENSDART00000164938 |
mapk15
|
mitogen-activated protein kinase 15 |
| chr23_-_28766019 | 1.56 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr11_+_14373731 | 1.55 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr14_+_11151485 | 1.54 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr9_+_17779858 | 1.53 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr21_+_22792068 | 1.52 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr8_+_18584936 | 1.52 |
ENSDART00000089161
|
ENSDARG00000062021
|
ENSDARG00000062021 |
| chr17_-_29102320 | 1.50 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr5_-_7901538 | 1.50 |
ENSDART00000159718
|
spef2
|
sperm flagellar 2 |
| chr23_-_20026717 | 1.49 |
ENSDART00000153828
|
atp2b3b
|
ATPase, Ca++ transporting, plasma membrane 3b |
| chr14_-_2728708 | 1.48 |
ENSDART00000031211
|
bicc2
|
bicaudal C homolog 2 |
| chr25_+_34340139 | 1.47 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr3_-_15475774 | 1.47 |
ENSDART00000136890
|
BX682550.1
|
ENSDARG00000090026 |
| chr6_+_102073 | 1.46 |
ENSDART00000151209
|
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr22_-_2921219 | 1.45 |
ENSDART00000092991
|
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr5_-_56981925 | 1.45 |
|
|
|
| chr13_-_6089739 | 1.44 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr20_-_47680305 | 1.43 |
ENSDART00000023058
|
efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr24_+_33697332 | 1.43 |
|
|
|
| chr8_+_50499355 | 1.43 |
|
|
|
| chr19_+_1106964 | 1.42 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr13_+_49527181 | 1.40 |
ENSDART00000176643
|
CABZ01084653.1
|
ENSDARG00000106801 |
| chr25_-_14573221 | 1.40 |
|
|
|
| chr17_-_53234493 | 1.39 |
|
|
|
| chr4_-_19027117 | 1.38 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr6_+_15635738 | 1.38 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr13_+_11305781 | 1.36 |
|
|
|
| chr10_+_9576710 | 1.36 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr12_+_6065116 | 1.36 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr2_+_54218444 | 1.34 |
ENSDART00000161221
|
capsla
|
calcyphosine-like a |
| chr20_+_52653111 | 1.34 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr21_+_17920229 | 1.33 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr6_+_29297594 | 1.32 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr25_-_3521512 | 1.31 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr4_+_5515028 | 1.31 |
ENSDART00000027304
|
mapk12b
|
mitogen-activated protein kinase 12b |
| KN149841v1_+_6400 | 1.31 |
ENSDART00000179534
|
CABZ01103873.1
|
ENSDARG00000108002 |
| chr23_-_2027166 | 1.28 |
ENSDART00000127443
|
prdm5
|
PR domain containing 5 |
| chr25_-_211879 | 1.28 |
ENSDART00000021614
|
srpk2
|
SRSF protein kinase 2 |
| chr20_+_40869520 | 1.27 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr24_-_37792982 | 1.25 |
ENSDART00000078828
ENSDART00000131342 |
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr13_-_31340029 | 1.25 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr1_+_2074153 | 1.24 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr23_+_22408855 | 1.24 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr21_-_32403403 | 1.24 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr10_-_29858525 | 1.24 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr3_-_22699157 | 1.24 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr2_-_2456819 | 1.23 |
ENSDART00000013794
ENSDART00000082155 ENSDART00000169871 ENSDART00000167202 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr21_-_28883441 | 1.22 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr1_-_30546026 | 1.21 |
ENSDART00000085370
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr4_+_76581339 | 1.21 |
ENSDART00000174203
|
zgc:113921
|
zgc:113921 |
| chr6_+_48138737 | 1.21 |
|
|
|
| chr1_+_2154115 | 1.21 |
ENSDART00000141036
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr10_+_6009364 | 1.20 |
ENSDART00000163680
|
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.3 | 6.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.3 | 7.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.1 | 3.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.0 | 4.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 1.0 | 5.8 | GO:0060295 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.9 | 2.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.9 | 2.7 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.9 | 3.5 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.8 | 2.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.8 | 2.3 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.7 | 2.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.6 | 3.2 | GO:0090317 | negative regulation of intracellular protein transport(GO:0090317) |
| 0.5 | 2.6 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.5 | 2.5 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.5 | 2.5 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.5 | 2.9 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.5 | 2.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.4 | 3.1 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.4 | 3.5 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.4 | 4.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.4 | 2.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 7.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 4.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.3 | 1.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 1.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 1.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 10.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 1.8 | GO:0030317 | sperm motility(GO:0030317) |
| 0.3 | 1.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.3 | 1.8 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 0.9 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.3 | 0.8 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.2 | 0.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 0.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 3.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.6 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.1 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.2 | 0.5 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 4.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 1.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 1.8 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 2.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 2.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.3 | GO:0006555 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.1 | 2.1 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 2.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.5 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.5 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.1 | 0.8 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 1.8 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.1 | 2.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 2.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.1 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 2.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 6.0 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.1 | 3.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 1.7 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.1 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.8 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 1.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.2 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 4.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.2 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 0.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 1.9 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 3.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.3 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.3 | GO:0043279 | response to nicotine(GO:0035094) response to alkaloid(GO:0043279) |
| 0.0 | 1.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 1.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.6 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.6 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 2.1 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 1.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 1.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 11.7 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 1.3 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 7.6 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 2.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.5 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 2.6 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 1.5 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0019320 | hexose catabolic process(GO:0019320) galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.3 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.9 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 0.7 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.8 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 1.0 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 2.3 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.7 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 2.0 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.8 | 4.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.6 | 2.5 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.6 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 4.0 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.6 | 2.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.5 | 2.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 6.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.4 | 2.7 | GO:0044447 | axoneme part(GO:0044447) |
| 0.3 | 3.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 3.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 3.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 4.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.9 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.2 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 7.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 20.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 3.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 3.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 9.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.0 | GO:0045111 | intermediate filament(GO:0005882) intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 11.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 10.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 10.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0001726 | ruffle(GO:0001726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.9 | 2.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.9 | 3.5 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.9 | 8.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 10.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 1.6 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 3.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.4 | 1.2 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.4 | 5.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 1.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 4.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 3.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 1.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 3.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 3.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.3 | 1.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 5.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 3.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 2.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 10.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 1.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 5.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.2 | 4.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 7.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.1 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 6.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 4.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 2.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 14.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 24.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 3.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 6.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 10.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 1.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 4.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.5 | 7.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 6.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 3.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 4.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 2.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 5.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.7 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |